Abstract
Ras-GTPase-activating protein (SH3 domain)-binding proteins (G3BPs, also known as Rasputin) are a family of RNA binding proteins that regulate gene expression in response to environmental stresses by controlling mRNA stability and translation. G3BPs appear to facilitate this activity through their role in stress granules for which they are considered a core component, however, it should be noted that not all stress granules contain G3BPs and this appears to be contextual depending on the environmental stress and the cell type. Although the role of G3BPs in stress granules appears to be one of its major roles, data also strongly suggests that they interact with mRNAs outside of stress granules to regulate gene expression. G3BPs have been implicated in several diseases including cancer progression, invasion, and metastasis as well as virus survival. There is now a body of evidence that suggests targeting of G3BPs could be explored as a form of cancer therapeutic. This review discusses the important discoveries and advancements made in the field of G3BPs biology over the last two decades including their roles in RNA stability, translational control of cellular transcripts, stress granule formation, cancer progression and its interactions with viruses during infection. An emerging theme for G3BPs is their ability to regulate gene expression in response to environmental stimuli, disease progression and virus infection making it an intriguing target for disease therapies.
Abbreviations: G3BP, Ras-GTPase-activating protein (SH3 domain)-binding proteins; ISGs, interferon stimulated genes; IFN, interferon; IFITM, interferon induced transmembrane protein; SG, stress granules; Ras-GAP, RasGTPase activating protein; RBPs, RNA binding proteins; EMT, epithelial to mesenchymal metastasis; MMP2, metalloproteinase protein 2; PTEN, phosphatase and tensin homolog; PMP22, peripheral myelin protein 22; BART, binding partner of ARL2; nsP3, non-structural protein 3; NTF2, nuclear transport factor 2; Ctnnb1, β-catenin; hnRNP, heterogeneous ribonucleoprotein
Keywords: G3BP, Rasputin, Stress granules, Translational regulation, Viral infection, Cancer biomarker
Graphical abstract
Highlights
-
•
Triage of many cellular mRNA occurs via stress granules in a G3BP-dependant manner.
-
•
G3BPs control intra cellular responses to viral infection.
-
•
Transcript stability, degradation and translation are controlled by G3BPs.
-
•
G3BPs can control cancer progression.
1. Introduction
G3BPs (also known as Rasputin (Rin) in Drosophila) were last reviewed in 2004 only 5 years after their initial identification and preliminary characterisation [1]. More than a decade later we are starting to make sense of their cellular activities and to tame their apparent biological promiscuity. Current consensus would undoubtedly agree that their central role is the triage of mRNA including translational control and RNA stability but this is being extended to other biological activities. However, research is still providing conflicting evidence regarding their precise role in these biological functions and would suggest that G3BPs sit at the nexus of several cellular functions and that they are involved in disease etiology including neurological disease, cancer progression and viral infection and these functions appear to revolve around G3BPs' roles in mRNA regulation and stress granule (SG) formation.
Ras-GTPase-activating protein (SH3 domain)-binding proteins derive their name from their original discovery which identified G3BP1 binding to the Ras-GTPase activating protein (RasGAP) [2]. The Ras, family of GTPases (which are key signalling transducers), in its active GTP-bound form, activates serine/threonine kinases such as Raf and initiates downstream signalling. Hydrolysis of the Ras-bound GTP molecule to GDP by RasGAPs, inactivates Ras which inhibits further signalling [3]. G3BPs were originally speculated to intersect with the Ras signalling pathway by interacting with the SH3 domain of RasGAP [2,4,5], but a recent study failed to fully support these results and provides evidence against G3BP1 being a genuine RasGAP-binding partner [6]. However, further experimentation needs to be done before making a final conclusion about the interactions between G3BPs and RasGAP. Apart from the potential interactions with RasGAP, the G3BP family has demonstrated significant interactions with other signalling pathways and these are discussed, in relation to cellular or disease contexts, in this review.
1.1. Structural motifs of G3BPs
In mammals, the G3BP protein family comprises of three homologous proteins; G3BP1, G3BP2a and its splice variant G3BP2b. All G3BPs have four distinct motifs; a nuclear transport factor 2 (NTF2) like domain [7], acidic and proline-rich regions, an RNA recognition motif (RRM) [8] and arginine and glycine rich boxes (RGG box) [9]. The difference between the G3BP2 splice variants lies in the proline-rich region where G3BP2b lacks 33 residues [4], however, the biological significance of splicing to generate G3BP2b has not yet been characterised. The NTF2-like domain shares both structural and functional homology to the small NTF2 protein which is involved in nuclear transport via nuclear pores [10,11]. The role of the NTF2-like domain in nuclear transport is supported by the presence of both G3BP1 and G3BP2 in the nucleus during serum stimulation [12], however, another study using targeted mutations in G3BP2 has shown that the NTF2-like domain was more important for targeting G3BP2 to the nuclear envelope and not for actual nuclear translocation of G3BP2 [13]. Moreover, the NTF2-like domain has been shown to facilitate protein-protein interactions [4] and can also mediate the dimerization of G3BPs [14]. Interestingly, G3BPs can also bind to Ran [15] and it is possible that the NTF2 regions interact with Ran at the nuclear pore, although this is yet to be confirmed.
The central regions of G3BPs contain the acid-rich motif which appears to be unstructured in nature and similar motifs in other proteins are often associated with protein-protein interactions as seen in transcription factors. It is yet to be determined if this region has any correlation with the data that demonstrates G3BP1 associates with acetylated histone 3-associated transcriptionally active genomic DNA [12]. The central region of G3BPs also contains the proline-rich region (typically identified by PxxP motifs). These regions are also associated with protein interactions and in particular, the binding to aromatic amino acids in target SH3 domains [16]. G3BP1 has three PxxP motifs which might limit its capacity to interact with partner proteins [17], compared to G3BP2a and G3BP2b which have five and six PxxP motifs, respectively [4]. The RRM of G3BPs have two conserved sequences, RNP1 and RNP2, which interact with target RNA sequences of 2–8 nucleotides through a beta sheet binding platform comprised of four beta strands with structural integrity provided by two alpha helices. The overall structure creates a three-dimensional platform that binds RNA [4,8]. The RRM can also bind with other proteins which may affect its specificity for RNA interactions [18].
RGG boxes are comprised of closely located arginine-glycine-glycine clusters from where the motif derives its name. The RGG containing region has an undefined exposed structure due to the larger polar amino acids like arginine, which surrounds the glycine residues. This exposed structure influences interactions with proteins or RNA and facilitates post-transcriptional modifications. Methylation of arginine residues blocks hydrogen bonds (H-bonds) which are important for target binding and consequently may affect protein-mRNA interactions [19]. Arginine methylation of heterogeneous nuclear protein (hnRNP) has been shown to play a role in the identification of the RNA complexes for nuclear export [20], however, contrary results were observed in mammalian cells where arginine methylation led to decreased retention of nuclear hnRNP A2 [21]. Later findings have questioned the hypothesis that arginine methylation of hnRNP A/B proteins is responsible for their nuclear/cytoplasmic distribution [22]. Whereas in relation to G3BPs, methylation of G3BP1 at Arg433 in the RGG box regulates Ctnnb1 (β-catenin) mRNA in a Wnt-dependent manner [23] while the Wnt-dependent methylation of G3BP2 recruits methylated G3BP2 into the dishevelled protein 3 super molecular complexes (Dvl3-complexes). The Dvl3-G3BP2 complex later facilitates low density lipoprotein receptor 6 (LRP6) phosphorylation at Ser1490 through GSKβ3. Methylation of G3BP2 is also involved in the regulation of Ctnnb1 mRNA, but unlike G3BP1, G3BP2 downregulation leads to an upregulation of Ctnnb1 transcript levels, but not the protein [24]. The methylation status of G3BP1 is also involved in SGs formation where demethylation of G3BP1, primarily at Arg-447, regulates SGs assembly in response to oxidative stress induced by sodium arsenite [25]. PRMT1, 5 and 8 are involved in the methylation of G3BP1, whereas the methylation by PRMT8 is very weak. PRMT1 and 5 controlled methylation of G3BP1 occurs in the RGG domain at specific residues. PRMT1 methylates Arg-435 and Arg-477 whereas Arg-460 is methylated by PRMT5. Methylation of Arg-477 strongly inhibits SGs formation whereas Arg-435 and Arg-460 remain methylated during SGs formation. This suggest that a fine tuned mechanism exists for methylation of GB3P1 and this may occur within the SGs. PRMT5 does not enter SGs whereas PRMT1 does enter SGs and may promote Arg-447 methylation in situ, stimulating SGs disassembly by taking G3BP1 out of the structure. The demethylation of G3BP1 to form SGs is functionally linked to JMJD6 (Jumonji C domain-containing protein 6), which is a component of SGs and interacts with G3BP1. JMJD6 is responsible for the direct or indirect demethylation of G3BP1 at three Arg-residues as its knockdown caused demethylation of G3BP1 and repressed SGs and these effects can be rescued by overexpressing JMJD6 [26]. More recently, it has been found that the LRP6, which is a canonical Wnt-receptor, inhibits Arg methylation of many proteins, including G3BP1 [27]. LRP6 inhibits the methylation of G3BP1 as demonstrated in LRP6 deficient mice which showed an increase (up to >30-fold) in the monomethylation (MMA) of the G3BP1 C-terminal domain [27]. In addition to SGs regulation, methylation of RGG region may affect the ATP/Mg2+ dependent DNA/RNA helicase activity of G3BPs [28].
1.2. Expression of G3BPs and their activity
Although G3BPs are expressed in all normal cells, some isoform specific tissue expression has been identified for G3BP1 in lung and kidney, for G3BP2a in brain and for G3BP2b in the small intestine [4]. G3BPs are primarily cytoplasmic proteins, but a difference in distribution has been reported for the different isoforms. G3BP1 can localize to nuclei in quiescent cells, most probably due to phosphorylation at Ser149 [5,29], however G3BP2 can move to nucleus upon serum-stimulation [11].
Earlier studies suggested that G3BPs could have a role in tumorigenesis as they are overexpressed in many cancers and proliferating cells [11,12,30]. G3BP1 has been found to be highly expressed in proliferating retinal pigment epithelial cells [31]. Furthermore, the generation of G3BP1 knockout (KO) mice supported the role of G3BP1 in cell survival and proliferation [32]. Moreover, G3BP1 (but not G3BP2) is involved in the cellular proliferation of various breast cancer cell lines through a regulatory effect on peripheral myelin protein 22 (PMP22) [33].
Although G3BPs possess a role in cell proliferation, it appears unlikely that G3BP proteins play a single specific function within the cell. Instead, emerging evidence suggests that these proteins may mediate alterations in gene expression at various levels of control, in response to a range of cell signals, in various cellular and sub-cellular contexts. For example, G3BPs form SGs [14], possess antiviral activities [34] and have a role in epithelial-to-mesenchymal transition (EMT)-induced metastasis [35,36]. The emerging roles of G3BPs in SGs formation, cancer metastasis and viral infection, has driven extensive research to explore the role of G3BPs in different cellular contexts. The following sections collate the latest articles describing the emerging roles of G3BPs with a view to understand and to update our knowledge about the dynamic roles of G3BPs and their involvement in cellular mechanisms in different cellular environments.
2. G3BPs in cancer
G3BP1 and G3BP2 are overexpressed in several human cancers particularly in breast [11,12,30], pancreas [37], colon, head and neck and lung cancers [11,30]. G3BPs are involved in various growth related signalling pathways which are involved in cancer progression such as Ras signalling, the NF-κB pathway, Erk signalling and the ubiquitin proteasome system [5,13,38] (see Table 1 ).
Table 1.
Summary of different cellular functions of G3BPs.
| Biological function | Description | Effect(s) | Reference(s) |
|---|---|---|---|
| Cancer etiology | G3BPs are overexpressed in various cancers | Promotes S-phase entry. Involved in Ras, NFкB and ubiquitin proteasome pathway signalling pathways. |
[11,30] [5,13,38] |
| G3BPs negatively regulates expression of p53 | Regulates tumour suppressing role of p53. | [42] | |
| G3BP1 is involved in Smad pathway | Facilitate cancer metastasis and invasion. | [36] | |
| G3BP1 regulates expression of PMP22 and BART mRNA. | G3BP1 supports cell proliferation and invasion through regulation of these transcripts. | [33,37,51] | |
| G3BP2 interacts with TWIST1. | G3BP2 regulates cellular localization of Twist1, having a role in cancer metastasis. | [35] | |
| G3BP1 interactions with mRNA | c-Myc mRNA | Degrades the c-Myc transcript by exonuclease activity & initiates RNA turn over. | [5] |
| Tau mRNA | Stabilizes Tau mRNA and affects neuronal differentiation. | [62] | |
| BART mRNA | Degrades BART mRNA and promotes metastasis and invasion. | [37,51] | |
| CTNNB1 mRNA | Degrades CTNNB1 mRNA thereby regulating the Wnt/β-catenin pathway. | [23] | |
| PMP22 mRNA | Degrades PMP22 mRNA and affects cell growth & proliferation. | [33] | |
| GAS5 & IGF-II mRNA | Degrades these transcripts by exonuclease activity. | [32] | |
| β-F1ATPase mRNA | Inhibits translation of β-F1ATPase. | [64] | |
| Grm5 mRNA | Inhibits expression of the mature Grm5 transcript. | [69] | |
| miR-15b ~ 16-2, miR-23a ~ 24a ~ 24-2 | Inhibits the maturation of both transcripts. | [69] | |
| miR-1 | Inhibits expression of mature miR-1 transcript | [67] | |
| HIV-1 RNA | G3BP1 sequesters viral RNA and inhibits the translation and protein packaging of viral proteins. | [116] | |
| G3BP2 interactions with mRNA | G3BP2 interacts with SART3 mRNA. | Stabilization of SART3 leading to expression of pluripotent transcription factors and breast cancer initiation. | [56] |
| G3BP1 and G3BP2 interactions with mRNA | Alphaviruses sfRNA interact with both G3BP1 & G3BP2. | Viral sfRNA sequesters G3BPs out of the SGs, hence compromising their antiviral activity. | [34,100] |
| Stress granule formation | G3BPs are essential part of mRNP complexes. | Recruit mRNP complexes to SGs. | [62,72] |
| G3BP overexpression induces SGs formation | Potentially stops translational initiation and affects many signalling pathways. | [14,48] | |
| G3BPs & viruses | G3BPs are sequestered out of SGs to viral foci. | nsP3 domain of alphaviruses target G3BPs to counteract the host's protective mechanisms. | [106,107,115] |
| 3C protease of poliovirus cleaves G3BPs. | Results in SGs inhibition and viral infection. | [119] | |
| G3BP1 interacts with IRES elements of FMDV & is cleaved by 3C protease. | Results in SGs inhibition and viral infection. | [121] | |
| G3BPs, along with Caprin1, regulate the IFN system during viral attack to reduce infection. | G3BPs are targeted by viral components to compromise the cell-based immune response to the virus. | [34] | |
| G3BPs are essential constituents of CHIKV and HCV viral fractions. | Facilitates viral replication and assembly. | [105,113] |
2.1. G3BPs and tumour suppressor genes
Although G3BPs are not oncogenes their expression levels are typically low in most of the normal cells whereas G3BPs expression levels are upregulated in many primary cancers and cancer lines. Increased expression of genes could be collateral damage caused by cancer progression, however, evidence would suggest that the regulation G3BPs is required by cancer cells and serve as auxiliary genes to promote the survival of cancer cells and this is best demonstrated through G3BPs' interactions with genes known to be directly involved in cancer progression. The tumour suppressor, PTEN, is mutated in numerous cancers and is implicated in pathways of proliferation and regulation of cell growth. PTEN regulates cellular activity by various mechanisms, the most important being through its phosphatase activity that specifically hydrolyses, and thus inactivates, the potent secondary messenger, phosphatidylinositol-3,4,5-triphosphate (PIP3) [39,40]. Furthermore, PTEN can modulate the expression levels of several proteins including G3BP1, AKAP121, DHFR, Rap1 and RCC1. It negatively regulates the protein expression levels of G3BP1 and AKAP121 [41]. To determine if the downregulation of G3BP1 protein by PTEN was due to the inhibition of signalling through PIP3, Huang et al. employed the use of a mutated PTEN (C124S, which cannot hydrolyse PIP3) and the inhibitors of PI3K (wortmannin and LY294002) to inhibit the formation of PIP3. Inhibition of PI3K reiterated the effect of PTEN downregulation, showing that PTEN suppresses the activity of G3BP1 through the PI3K pathway via its phosphatase activity [41]. Therefore, a potential pathway to tumorigenesis appears to be 1) through loss of functional mutations in PTEN, resulting in 2) overexpression of G3BP1, leading to 3) cellular proliferation and deregulated promotion of the cell cycle, possibly through its endonucleolytic degradation of growth arrest genes like GAS5, as previously described by Zekri et al., 2005 [32]. To summarize, the overexpression of G3BP1 displays a diseased state by facilitating cellular proliferation, apparently through regulation of the cell cycle and this seems to be linked with the ability of G3BP1 to associate with RNA.
G3BPs bind to tumour suppressor p53 both in vivo and in vitro and are involved in the redistribution of p53 from the nucleus to the cytoplasm. Moreover, G3BPs negatively regulate the expression of p53 because the knockdown of G3BP1 and G3BP2 by short hairpin RNA (shRNA) resulted in an upregulation of p53 in human cancer cell lines whereas their overexpression leads to a differential localization of p53 in cells [42]. G3BP2, but not G3BP1, also interacts with murine double minute 2 (MDM2), which is a negative regulator of p53 and exports p53 from the nucleus to the cytoplasm. The interaction between G3BP2 and MDM2 compromises its ability to ubiquitinate p53 and the subsequent degradation of p53 by the proteasome [42]. A cytoplasmic lncRNA, P53RRA, interacts with the RRM domain (also responsible for the interaction of G3BP1 with p53) of G3BP1 in the cytoplasm via nucleotides 1 and 871 and this interaction displaces p53 from G3BP1-complex, subsequently retaining p53 in the nucleus, inducing cell cycle arrest and cell death [43]. P53RRA also increases the levels of MDM2 and p21 (which is a direct target of p53), showing that P53RRA promotes the activity of the p53 signalling pathway [43]. Regardless of their mechanism of action, both G3BP proteins inhibit p53 functions and hence play an accessory role in tumorigenesis. Furthermore, G3BP2 is transcriptionally activated by the androgen receptor (AR) and facilitates the proliferation of prostate cancer [44]. Upon androgen-mediated induction, G3BP2 interacts with the SUMO-E3 ligase, RanBP2, which facilitates the AR-dependent sumoylation of p53 and regulates the translocation of p53 from nucleus to cytoplasm [45]. The strong cytoplasmic localization of p53 is clinically correlated with elevated G3BP2 expression and predicts poor prognosis and disease progression to the hormone-refractory state, suggesting a role of G3BP2 in cancer metastasis and progression [45]. More recently, it has been found that tripartite motif-containing protein 25 (TRIM25) interacts with G3BP2 and is responsible for modulating the cellular localization of p53. TRIM25 is involved in the negative regulation of p53 through an interaction with G3BP2 and its over-expression suppresses the p53 activity via a G3BP2-mediated export mechanism [46]. This study also suggests that TRIM25 could serve as a positive regulator of p53 sumoylation induced by a G3BP2/RanBP2 complex and a negative regulator of p53 ubiquitination induced by MDM2 [46]. Furthermore, USP10, a binding partner of G3BP2, is also an androgen responsive gene and is transcriptionally activated by the AR. USP10 regulates androgen-mediated signalling and inhibits p53 activities by regulating G3BP2 expression. The association of USP10 with G3BP2 suppresses p53 signalling which correlates to a poor prognosis in prostate cancer, highlighting an oncogenic role of USP10 in prostate cancer via G3BP2 [47]. These studies suggest a role of G3BPs, specifically G3BP2, in the mediating the tumour suppressor activities of p53 and thereby implicating it in cancer development or progression.
Interestingly, G3BP1 and G3BP2 play an important role in SGs formation [48] and this activity has also been associated with cancer progression. Various cellular stresses like hypoxia and heat shock induce SGs formation, however, of particular interest is the finding that chemotherapeutics may also induce SGs formation which may in turn inhibit some stress responsive pathways (like the MAPK pathway) and hence suppresses apoptosis [49], suggesting yet another role of G3BPs in cancer progression. In this case, through the regulation of the MAPK pathway [50].
2.2. G3BPs in cancer metastasis and invasion
G3BP1 binds to the transcript of BART (Binding partner of ADP-ribosylation factor-like 2 (ARL2)), and facilitates its degradation whereas CD24 interacts with G3BP1 and prevents its endoribonuclease activity on BART in SGs, preventing metastasis and cell invasion in pancreatic cancer cells [51]. The association of G3BP1 and BART is dominant-negatively inhibited by overexpression of the N-terminal domain of G3BP1, subsequently contributing to the posttranscriptional regulation of cell invasiveness and metastasis in pancreatic cancer cells [37]. G3BPs also facilitate the invasion and migration of the human lung cancer cells through activation of Src, FAK, ERK, NFκB with subsequent activation of matrix metalloprotease (MMP) 2, 9 and plasminogen activator (uPA). Both stable and transient downregulation of G3BPs suppressed metastasis and invasion of human lung cancer cells by suppressing the Src, FAK, ERK, NFкB and lowered levels of MMP2 [50]. Src and FAK kinase stimulate the MEK/ERK pathway which initiates a cascade of events, including activation of NFκB, leading to activation of factors which support cancer metastasis and invasion such as MMP2, MMP9 and uPA in lung cancer cells [50]. Therefore, the inhibition of Src and FAK by G3BPs knockdown suggests a role of G3BPs in human lung cancer metastasis. G3BP1 may also play a vital role in lymph node metastasis and invasiveness in oesophageal squamous cell (ESC) carcinoma as overexpression of G3BP1 is positively correlated with poor prognosis in patients with ESC. Although the modes of action through which G3BP1 can induce metastasis are still unclear, it can be considered as an independent marker for the prognosis of carcinogenesis in ESC patients [52]. Depletion of G3BP1 inhibits SGs assembly and cancer invasion in sarcoma xenografts, and entirely inhibits metastasis of lung cancer in mouse models [53]. Although no correlation has been observed, the analysis of G3BP1 and Vezatin (VEZT, a putative tumour suppressor) expression levels in gastric cancer demonstrates a downregulation of both G3BP1 and VEZT at mRNA levels. The significance of this was observed in relation to expression of these proteins with the age of patients and stage of disease. Expression levels of VEZT were directly related to metastasis whereas no significant relevance between G3BP1 expression and metastasis was reported. The conclusion of this research was that G3BP1 can be used as a diagnostic marker for gastric cancer whereas VEZT would be a preferred biomarker for gastric cancer progression [54]. The data regarding G3BPs and metastasis does not show clear correlations in all cancers and appears to be contextual in relation to the type and stage of the cancer and therefore needs more research to clarify the significance of the data reported so far, however, some of these correlations are discussed below.
2.3. G3BPs in EMT (epithelial to mesenchymal transition)-induced breast cancer metastasis
Several studies (as described above) have shown that G3BPs are overexpressed and are involved in metastasis and invasion by various cancers, but they have also been specifically implicated in the EMT-induced metastasis of breast cancer. Overexpression of G3BP1 mediates EMT in breast cancer cells via the Smad signalling pathway. siRNA mediated knockdown of Smads entirely blocked G3BP1-induced EMT, signifying the role of the Smad signalling pathway in this process. Likewise, G3BP1 knockdown blocked the mesenchymal phenotype of MDA-MB-231 cells in vitro and repressed tumour growth and metastasis in 4T1 cells in vivo, demonstrating that G3BP1 has a role in breast cancer progression and may serve as a potential therapeutic target for metastatic human breast cancer. G3BP1 plays an essential role in the activation of Smads through phosphorylation which in turn stimulate EMT factors [36]. Furthermore, G3BP1 is overexpressed in hepatocellular carcinoma (HCC) and is involved in EMT of HCC by stimulating the expression of Slug, a member of the SNAIL family of zinc finger transcription factors which induces EMT. Both in vitro and in vivo studies have shown that downregulation of G3BP1 decreased cell migration and metastasis. G3BP1 depletion reduced Slug expression and increased the expression of E-cadherin, subsequently inhibiting metastasis [55]. G3BP2, on other hand, inhibits matrix-stiffness induced-EMT in MCF10A (human) and Eph4Ras (mouse) cell lines by sequestering TWIST1 (an important mechano-mediator) into the cytoplasm. Matrix stiffness mediates EMT via the TWIST1-G3BP2 mechano-transduction pathway. During matrix stiffness, TWIST1 detaches from G3BP2 and moves to nucleus, resulting in EMT induction. Moreover, G3BP2 knockdown leads to constitutive localization of TWIST1 to the nucleus and subsequent induction of EMT, suggesting that the TWIST1-G3BP2 mechano-transduction pathway responds to biomechanical signals from the tumour microenvironment to drive EMT [35]. However, a more recent study contradicts these findings by suggesting that G3BP2 has a role in breast cancer initiation by stabilizing mRNA transcripts of squamous cell carcinoma antigen recognized by T cells 3 (SART3). SART3 is responsible for the expression of pluripotent transcription factors, Oct-4 (octamer binding protein 4) and Nanog. These findings support the role of G3BP2 as a breast cancer initiating protein. Moreover, this report suggests that a lead anticancer compound, C108, interacts with G3BP2 via its RRM thereby facilitating the degradation of SART3 mRNA with subsequent tumour suppression [56]. These reports suggest that G3BP2 can serve as a positive regulator of breast cancer initiation as well as a negative regulator of cancer metastasis. These two contradictory roles of G3BP2 can be due to the fact that to acquire cancer initiating properties for metastatic colonization the cancers cells must lose their EMT-phenotype [57].
The role of G3BPs in cancer progression and metastasis seems to be contextual and tumour type dependent and therefore, the exact mechanisms of their involvement are still ambiguous and need further exploration. To date the role of G3BP1 in cancer progression has been studied more than G3BP2 and it appears that G3BP1 participates in metastasis and invasion of various cancers including breast cancer, HCC, ESCC, lung cancer and oesophageal cancer but the precise pathways that are activated by G3BP1 are still ambiguous because a single consensus mechanism has not been identified. The current data is stochastic because the various discoveries of G3BP1 in cancer progression have been identified by its participation in cancer related pathways. Perhaps the role of G3BPs would be more easily identified if a systematic approach to determine the functions of the protein had been carried out. The current discoveries are almost serendipitous and have implicated G3BP1 in the regulation of tumour suppressor genes like p53, SGs formation and various signalling pathways like the Src/FAK and Smad signalling pathways. Although G3BP2 has been identified as being overexpressed in many cancers, its role is less characterised than G3BP1. Furthermore, G3BP2, appears to have contradictory roles in cancer progression, as it has been reported to inhibit as well as initiate breast cancer. Like G3BP1, G3BP2 is also involved in SGs formation and p53 regulation and its role in cancer progression could also be due to these activities. However, the role of G3BP2 has only been studied in breast cancer metastasis and therefore, there is a need to explore its role on other cancers before we come to final consensus about its role in cancer progression. Overall, these studies have shown significant roles of G3BPs in cancer metastasis and invasion, suggesting a potential for GB3Ps to be targeted as a putative drug target to suppress tumorigenesis.
2.4. G3BPs as potential drug target
A novel peptide, GAP161, binds with G3BPs and interferes with their interaction to RasGAP, thereby inhibiting Ras signalling, which in turn induces apoptosis and suppresses cell growth. GAP161 causes G3BP1 and G3BP2 downregulation and the knockdown studies of G3BPs have led to decreased levels of proliferation in HCT116 cells both in vivo and in vitro which indicates that targeting G3BPs might be useful in cancer treatment [58]. Moreover, another synthetic peptide, GAP159, targets and inhibits the expression of G3BPs thereby increasing CDDP-induced cytotoxicity in HCT116 cells and CT26 mouse models [59]. Another biologically active compound present in green tea, called Epigallocatechin gallate (EGCG), is known to inhibit lung cancer proliferation by interacting with G3BP1, subsequently inhibiting Ras and other downstream signalling pathways such as the mitogen activated protein kinase (MAPK) pathways [60]. Resveratrol, an anticancer agent, targets G3BP1 by binding to its NTF2-like domain thereby inducing apoptosis by p53 activation. Moreover, downregulation of G3BP1 suppresses resveratrol induced p53 expression and apoptosis [61]. Altogether, these lines of evidence suggest that G3BPs are involved in cancer progression and metastasis in through several pathways (Fig. 1 ) and can serve as a potential drug target for future studies. However, it is important to bear in mind that neuronal cell death and embryonic lethality has been detected in G3BPs-deficient mice, therefore, systemic therapies that modulate G3BPs activity may not be an appropriate approach as a therapeutic. This suggests that targeting specific tissues or cancers cells may be required if G3BPs are to be targeted in anti-cancer therapies and this would represent a significant hurdle if drugs targeting G3BPs are to be considered. Furthermore, the essential roles for G3BPs in regulating gene expression during cancer development are still to be fully characterised. The data currently available for G3BPs highlights the need for further studies to explore the potential for targeting G3BPs as a cancer therapeutic.
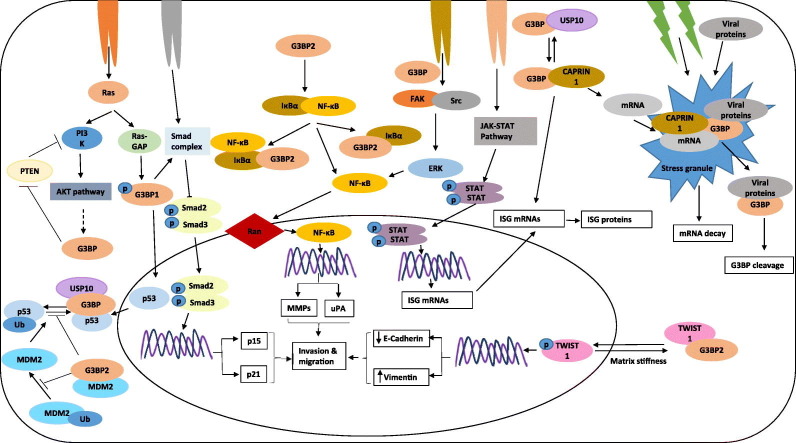
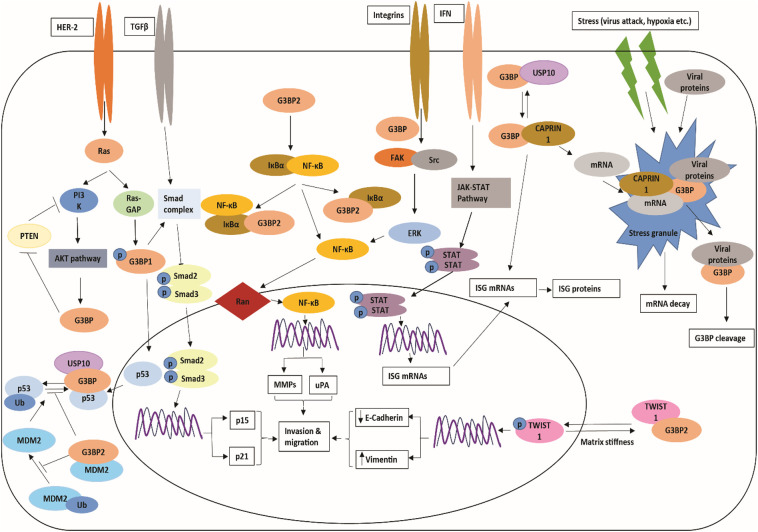
Fig. 1.
Schematic representation of G3BPs in different cellular pathways.
3. G3BPs and RNA interactions
3.1. G3BPs and mRNAs
Functional studies of G3BPs have extensively been focused on finding their mRNA targets, however, to date only one mRNA target, SART3 (as mentioned above), has been identified for G3BP2. RNA immunoprecipitation (RIP) and qPCR analysis of SART3 mRNA in G3BP2 depleted cells as compared to control cells revealed that SART3 mRNA degrades in G3BP2 depleted cells [56]. On the other hand, many RNA targets have been identified for G3BP1. The first reported interaction for G3BP1 was with c-MYC mRNA and G3BP1 exhibited endonuclease activity on this target in a phospho-dependant manner in vitro, subsequently cleaving the 3′-UTR of the c-MYC mRNA [5,29], however, in vivo analysis did not support these findings [32]. It is difficult to reconcile the discrepancy between the in vitro and in vivo data but it is possible that the in vivo analysis described by Zekri et al., was performed in G3BP1-KO (G3BP1−/−) fibroblasts. However, G3BP2 would still have been present in the cells and it is unclear if there are redundancies between the proteins which may also mediate the regulation of c-MYC mRNA. Therefore, it is possible that the in vivo study may not have had the capacity to quantify the endoribonuclease effect of GB3P1 on c-MYC without depleting G3BP2. Furthermore, the in vivo analysis may have been unable to evaluate short-lived mRNAs, such as the transcript of c-MYC, under these conditions [32]. Therefore, the interaction of G3BP1 with c-MYC transcript and its endoribonuclease action on this transcript is still ambiguous and needs further exploration before coming to a final conclusion.
Although the endoribonuclease activity of GB3P1 on c-MYC could not be completely characterised by Zekri et al., additional studies have supported this function of G3BP1 for other transcripts [29] and it can be applied to other mRNAs including; BART [37], CTNNB1 [23], PMP22 [33], IGF-II and GAS5 [32] (see Table 1). Conversely, G3BP1 has been found to play a role in stabilizing mRNAs including the transcripts of TAU [62] and CDK7 [63]. G3BP1 binds with the 3′-UTR of β-F1ATPase and impedes its translation, supporting a role of G3BP1 in the Warburg effect during cancer progression [64]. The regulation of a specific mRNA transcripts bound by G3BP1 may be highly regulated and depend on post-translational modifications of G3BP1 and its association with other proteins. For instance, the binding of G3BP1 with CDK7and CDK9 mRNA transcripts depends on the interaction of G3BP1 with RasGAP and filamin [63]. In its phosphorylated form, G3BP1 exhibits endoribonuclease activity while unphosphorylated G3BP1 appears to regulate cell proliferation and this suggests that the phosphorylation status of G3BP1 may function as a cell growth switch where phosphorylated G3BP1 mediates degradation of growth-related mRNAs and thus reduce cellular proliferation [1].
G3BP1-regulated mRNAs have been found to affect cell growth in both a positive (e.g. c-MYC [5] and CDK [63]) and negative manners (e.g. GAS5 [32]). The RNA binding specificity of G3BP1 has in several different cases been shown to be influenced by protein interactions. For example, CD24 interaction with G3BP1 can inhibit BART mRNA decay which leads to increased invasion capacities of pancreatic cancer cells [37] whereas interaction with Caprin1 affect its localization to SGs [65]. SGs are specialised ribonucleoprotein (RNP) particles that have a role in triaging and regulating the life cycle of RNA and are discussed further below (see section: G3BPs and stress granules (SGs) formation). Bidet et al., reported that Dengue virus (DENV) generate a subgenomic non-coding flaviviral RNA (sfRNA) from the 3′-UTR of viral genomic RNA (gRNA) which bind to host proteins including G3BP1, G3BP2 and Caprin1, which are involved in the translational regulation of ISGs and antagonize their antiviral activity [34].
The interplay between G3BPs, SGs and RNAs are not only limited to endogenous mRNAs but are also extended to microRNAs (miRNAs). These interactions will be discussed in more detail below.
3.2. G3BPs and microRNAs (miRNAs)
Apart from interactions with endogenous mRNA and viral RNA, G3BP1 is also involved in the processing of several miRNAs. For example, overexpression of G3BP1 in cardiac hypertrophy inhibits miR-1 processing, by interacting with its consensus sequence present in the pre-miR-1-2 stem loop which results in a subsequent increased expression of miR-1 targets (CDK9, eIF4E) which have a significant role in the development of cardiac hypertrophy. Although the overexpression of G3BP1 alone is not enough to cause development of cardiac hypertrophy, it does have a role in the regulation of miR-1 during disease progression [67]. The potential role of G3BP2 in miRNA regulation in cardiac hypertrophy remains elusive, but it is involved in isoproterenol-induced cardiac hypertrophy by inducing the NFκB signalling pathway [68]. Interestingly, in situ hybridisation studies of G3BP2 mRNA in developing mouse embryos showed that its expression is reduced in the heart relative to other tissues (unpublished data by the author).
G3BP1 also regulates the maturation of the glucocorticoid receptor (GR) regulated miRNAs. It binds to primary miR-15b ~ 16-2 and miR-23a ~ 27a ~ 24-2 through its consensus sequence and restricts their maturation in endothelial cells [69]. G3BP1 is also involved in the regulation of neuronal genes by regulating non-coding (NC) RNAs, including intron retaining transcripts. The immunopurification of G3BP1 complexes from the mouse brain has shown that it primarily interacts with the RNA transcripts retaining introns or with NC-RNA sequences like 3′-UTRs and long NC-RNAs. G3BP1 specifically inhibits the expression of mature glutamate receptor 5 (Grm5) RNA and this appears to be regulated by binding with introns present in the premature transcript to stabilize the premature transcript in the cerebellum [70]. No miRNA target for G3BP2 has been reported yet, however, it has been reported that miRNA-23b targets G3BP2 during diabetic neuropathy to increase albuminuria and fibrosis. High blood glucose levels decrease miR-23b levels whereas the expression of G3BP2 (a putative target of miR-23b) is increased in kidney cells. Moreover, in vitro studies showed that G3BP2 levels are decreased by overexpression of miR-23b, and increased by inhibition of miR-23b levels and this miR-23b/G3BP2 circuit is important in regulating the pathogenic pathways during diabetic neuropathy [71].
In conclusion, the role of GB3P1 in RNA metabolism has been characterised more than that of GB3P2. G3BP1 is involved in the regulation of various cellular RNAs, including mRNAs and miRNAs. The role of G3BP1 in mRNA metabolism is, once again, contextual in nature with the data showing that specific transcripts are controlled differently depending on the cell and the stimulus. It has both stabilizing (TAU, CDK7) and degrading (c-MYC, BART, CTNNB1, PMP22) effects on its target mRNAs. In addition, it also inhibits (βF1ATPase) and enhances (ISGs) the translation of various target mRNAs. The control mechanisms which allow GB3P1 to differentially regulate its targets have not been clearly identified. The variations in control demonstrated by G3BP1 could be cell type or stimuli specific and may depend on the cis-factors present in its mRNA targets. Possibly, G3BP1 recognizes multiple sequences or elements in different targets and the fate of the transcript is determined by the RNA elements to which G3BP1 binds. However, we cannot exclude the possibility that the fate of a transcript is determined by several trans-factors working in conjunction with G3BP1. Therefore, there is need to catalogue the RNA sequences recognized by G3BP1 and determine how GB3P1 uses these to control the fate of the transcript. Several miRNA targets have also been reported for G3BP1 and so far it appears that for miRNAs G3BP1 has one mode of action and that is to stop the maturation of target pre-miRNAs by binding to them and this control mechanism appears to be extended to NC-RNAs as well. Despite the huge sequence homology shared with G3BP1, G3BP2 does not appear to have a complicated role in RNA metabolism and to date there is only one reported mRNA target, SART3, for G3BP2. However, recently G3BP1, G3BP2 along with Caprin1 have reported to have role in the translation of ISGs. To date the role of G3BPs in the regulation of RNA metabolism is not well-characterised and there are many unknowns, including consensus sequences for the RNA elements bound by G3BP1 and GB3P2. These elements need to be identified before we can understand how G3BPs regulate the fates of RNAs. Future studies should focus on identifying the G3BPs' RNA-binding elements and although in silica strategies have been made using bioinformatics analysis, those preliminary studies will undoubtedly become more powerful as in vitro studies identify and confirm validated targets.
4. G3BPs and stress granules (SGs) formation
The functions of G3BP proteins differ according to cellular context, however, the functions are generally attributed to cell proliferation and survival. Both G3BP1 and G3BP2 interact with polysome-associated mRNP complexes [62,72] and the key role for this might be to regulate translation initiation of mRNAs and/or induce SGs formation.
SGs are translationally stalled mRNP complexes, formed in the cytoplasm in response to various cellular stresses such as oxidative stress, hypoxia and viral infections (as reviewed by [[73], [74], [75]]). The overall role of the SGs is to triage mRNA [76] although a role of SGs in protein sequestration cannot be excluded because studies have shown that SGs also recruit proteins which are involved in signalling pathways and hence influence cell metabolism and survival [77,78]. The dynamic transport of mRNPs between translating polysomes and translationally silent bodies such as SGs is a result of mRNA sorting. Exposure to stressful conditions leads to SGs assembly [79], but both the extent and form of stress may affect the components of SGs. Eukaryotic cells shut down some cellular translation in response to environmental stresses (common environmental stresses are hyperosmolarity, heat and oxidative conditions), in order to save energy and to respond with stress induced damage [79]. The assembly of canonical SGs depends upon the phosphorylation of eIF2α and different stresses induce the specific serine/threonine kinases which phosphorylate the eukaryotic initiation factor 2α (eIF2α), including HRI (heme-regulated initiation factor 2α kinase) which senses the oxidative stress induced by sodium arsenite [[80], [81], [82]], PKR (protein kinase RNA-dependent kinase) which is activated by heat shock, viral infections and UV irradiation [74,81,82], PKR-like endoplasmic reticulum (ER) kinase (PERK), induced in ER lumen by unfolded proteins and general control non-derepressible 2 (GCN2) protein which is activated by stress induced by amino acid deprivation [81,82]. Upon specific stress stimuli, these kinases phosphorylate eIF2α at Ser51 leading to the depletion of the eIF2/tRNAi Met/GTP ternary complex which is responsible for translational initiation. In the absence of the ternary complex, formation of the 48S preinitiation complex, which assembles at the 5′-ends of capped mRNAs, is impaired producing a translationally stalled, noncanonical 48S complexes which are unable to recruit the 60S ribosomal subunit [83,84] and these non-functional translational initiation complexes aggregate to form SGs [85]. Alternatively, translational inhibition caused by suppressing the functions of eIF4A and eIF4G also induces SGs formation [85], but these pathways are not discussed in detail here as are outside the scope of this review.
During stress, both G3BP1 and G3BP2 localize in eIF2α-induced SGs [14] but they also harbor the potential to induce SGs independently [48]. G3BPs are recruited to SGs in an unphosphorylation dependent manner suggesting that the phosphorylation status of G3BPs might influence the fate of the mRNAs by protecting it from degradation during cellular stress. Under normal conditions, G3BPs are phosphorylated and in some reported cases, this causes mRNA degradation, whereas upon cellular stress induced by arsenite, unphosphorylated G3BPs may oligomerize and bring mRNAs to the SGs. In concordance with this, arsenite leads to unphosphorylation of G3BPs at Ser149 with subsequent SGs formation both in mammalian cells [14,86] and in Drosophila cells [87]. Whereas, upon the stress induced by amino acid starvation in Drosophila S2 cells, only the Ser142 phosphorylated Rasputin (G3BP) is recruited to SGs [87]. Furthermore, Sec16 which is a component of endoplasmic reticulum (ER) exit site, interacts with and stabilizes phosphorylated Rasputin. However, in absence of Sec16, the stabilization of phosphorylated Rasputin is not sufficient to form SGs, suggesting a critical role of Sec16 in SGs assembly upon amino acid starvation [87]. These studies suggest that SGs formation is a fine-tuned process which is regulated by specific signals that are unique to each stress.
Besides stress inducing agents, overexpression of several RBPs such as TIA-1, CPEB1 and G3BPs along with inhibition of translational initiation complex components [88,89] can induce SGs assembly [14,90,91]. Most stress factors suppress translational initiation, forming SGs by inducing phosphorylation of the eIF2α whereas SGs induction by G3BPs are independent of this phosphorylation and could affect the translational complex afterwards by PKR-dependent phosphorylation of eIF2α [92]. However, another study has shown that the overexpression of the C-terminal region of G3BPs can cause phosphorylation of eIF2α [37]. Therefore, it remains to be elucidated if the SGs induction by G3BPs is dependent or independent of eIF2α phosphorylation or if the specific cellular contexts decide this dependence. In brief, G3BPs induce SGs assembly and co-localize with another SGs marker protein TIA-1 [14,92,93] however, it is yet to be confirmed whether these two mechanisms are entirely dependent on one another.
Interaction of G3BPs' own transcript and protein with other proteins also affects its ability to form SGs during various cellular stress responses. TAR DNA-binding protein 43 (TDP-43) regulates SGs through differential regulation of G3BP1 and TIA-1 by regulating the aggregation of TIA-1 and the mRNA levels of G3BP1. Depletion of TDP-43 leads to disrupted aggregation of TIA-1 and also downregulates the mRNA levels of G3BP1, though it should be noted that this study does not show the direct interaction of the TDP-43 with G3BP1 mRNA [94]. In addition, another protein, YB-1, has been reported to regulate the expression levels of G3BP1 through its interaction with the 5′-UTR of G3BP1, subsequently regulating SGs formation during stress [94]. Caprin1 interacts with G3BP1, forming a complex which is localized in the SGs though the association of G3BP1 and Caprin1 is not mandatory for their ability to form or to be sequestered into SGs [65]. However, a recent study suggests that G3BP proteins interact with Caprin1 and USP10 but that they are mutually exclusive allowing the binding of these proteins to serve as a switch for SG formation depending on the binding partner interacting with G3BP1. When the interaction of G3BPs is with Caprin1 the complex facilitates SG formation, whereas their binding with USP10 inhibits SGs assembly [86]. Earlier reports have shown that knockdown of G3BPs suppress SGs formation [95] whereas this report showed that G3BPs depletion abrogates SGs formation only in response to stresses that act through the phosphorylation of eIF2α or to the inhibition of eIF4A and the cells lacking G3BPs are still competent to form SGs in response to non-eIF2α or eIF4α dependant pathways such as osmotic or thermal stress. Rescue experiments using a G3BP1 mutant that lacks the ability to bind Caprin1 or USP10 rescues SGs assembly, whereas other phosphomimetic mutants of G3BP1 (G3BP1-S149E) fail to do that [86]. G3BP2 is a binding partner of protein kinase C (PKCα), which also has a role in SGs formation [96] and this interaction might assist G3BP2 in SGs assembly. A ubiquitously expressed protein, Tudor-SN interacts with G3BP1 and is co-localized with G3BP1 in SGs. This interaction is not important for SGs assembly, however, the depletion of Tudor-SN affects SGs aggregation [97].
G3BP1 and G3BP2 are both considered as basic components of SGs and can induce the formation of SGs, however, their exact role in SGs formation is still not clearly defined. Early reports suggested that upon stress stimulation G3BPs are unphosphorylated which acts a signal to recruit them to SGs but more recent studies have shown that the type of stress decides whether phosphorylated or unphosphorylated G3BPs are recruited to SGs. How these decisions are made, based on the different phosphorylation states of G3BPs, is still not known but this might be due to the differential binding partners of G3BPs under different stress stimuli and therefore, there is a need to further explore and characterise the role of G3BPs under different cellular stresses. In conclusion, the role of G3BPs in SGs is associated with the triage of mRNA under environmental stresses but like other activities of G3BPs, this activity also appears to be target and context specific and needs further exploration.
5. G3BPs in viral infections
SGs are also induced in response to viral infections [98], most likely as a cellular response to block virus replication of survival, however, several viruses have evolved different counteractive mechanisms to avoid negative regulation by SGs and many of these target G3BP proteins [99,100]. For instance, old and new world alphaviruses exploit various essential components of SGs, like G3BPs and FXR [100], probably to inhibit the formation of SGs in response to viral stress. The sequestration of essential SGs components, such as G3BPs, by viral components to foci outside and exclusive to SGs may give additional clues to the role of G3BPs and these interactions will be discussed below.
The precise role of G3BPs during viral infections has yet to be fully elucidated and in an apparent contradiction to the inhibitory role of G3BPs mentioned above, it appears that in some instances G3BPs are utilised by viruses to support their replication or the transcription of viral genes which seems contradictory to the role of G3BPs to induce the innate immune system in response to viral infection. Viruses manipulate and hijack cellular processes to favour conditions for their survival so it is of no surprise that viruses may use the SGs components like G3BP1 and TIA-1 to favour viral replication and/or to invade the cellular immune response. In this section the different ways by which viruses manipulate G3BPs to gain control of host cellular system are discussed.
5.1. G3BPs and viral transcription
Initial reports indicated that G3BP1 has a role in viral transcription by regulating the activities of vaccinia virus RNA polymerase and transcription factors [101] and in regulating hepatitis C virus (HCV) virus replication [[102], [103], [104]]. G3BP1 has been reported to play an important role in the viral assembly of HCV, as it is a constituent of the viral replication complex (RC) [105]. G3BP1 has been shown to interact with the NS5B protein of HCV and HCV RNA [103,104] and later G3BP1 was confirmed to be the part of the RC of HCV [102]. The role of G3BP1 in HCV replication was confirmed by downregulating G3BP1 in viral infected cells which resulted in a significant reduction of viral particles, confirming a role of G3BP1 in viral replication [102].
5.2. Viral targeting of G3BPs by protein interactions
Despite the findings implicating G3BP1 in HCV replication, G3BPs are now emerging as antiviral proteins in other systems and research has shown that many viruses use distinct mechanisms to target G3BPs in order to invade a cell based immune-like response. SGs are formed in mammalian cells in response to environmental stress including viral infections by various viruses [74]. Both G3BP1 and G3BP2 are sequestered to the foci which contain aggregations of viral proteins and this is facilitated by the C-terminal variable repeat domains of nsP3 (non-structural protein 3) proteins of old world alphaviruses which bind directly to G3BPs [106,107]. The nsP3 interactome studies of Sindbis virus (SINV) show that both G3BP1 and G3BP2 are present in the nsP3 containing complexes. Even Rasputin (G3BP1 homolog in insects) is associated with nsP3 containing complexes [108]. The nsP3 C-terminal variable domain of many alphaviruses contains an SH3-domain binding motif (PxxPxR) which facilitates its interaction with G3BPs, resulting in G3BP's recruitment to viral cytoplasmic foci and subsequent inhibition of SGs assembly [109]. The interaction of nsP3 with G3BPs has also been shown in CHIKV (chikungunya) infection. G3BPs' NTF2-like domain has been reported to interact with the nsP3 protein of many viruses, including Semliki Forest virus (SFV), SINV, CHIKV and Herpes Simplex virus [107,[110], [111], [112], [113]].
CHIKV infection mediates cytoplasmic G3BP1 and G3BP2 containing granules which differ from the actual SGs in terms of morphology and behaviour. Although knockdown of G3BPs has shown that G3BPs play an essential role in efficient viral replication, it is assumed that they do not have any direct role in RNA synthesis as the cellular fractions containing CHIKV replication/transcription complexes do not contain G3BPs [113]. Moreover, the study of the interaction of CHIKV nsP3 protein and Rin (G3BP) in insect cells and live mosquitos (Ae. Albopictus) shows that the normally diffuse cytoplasmic localization of Rin, is effectively sequestered to granules, containing nsP3, during co-expression studies. The interaction of Rin and nsP3 is moderated through an interaction of the C-terminal domain of nsP3 and the NTF2-like domain of G3BPs [114]. More recently it has been shown that viral and cellular proteins containing FGDF motifs also bind with G3BPs and inhibit SGs assembly by G3BPs. The nsP3 protein of SFV contains two FGDF motifs while the ICP8 protein of HSV contains one FGDF motif and these motif(s) are responsible for their binding to G3BPs. Interestingly, the binding interaction of USP10 with G3BPs is also mediated by a FGDF motif [115]. The N-terminal Capsid domain of Gag (precursor viral polyprotein, processed into different viral proteins during maturation) interacts with eukaryotic elongation factor 2 (eEF2) to block SGs formation. To dismantle the already formed SGs, Gag recruits G3BP1 out the SGs (by displacing eEF2 with G3BP1) [116]. Infection with Rubella virus has also been shown to change the localization of host G3BPs, indicating that G3BPs may play an important role during infection by Rubella [117].
5.3. Plant viruses and G3BPs
The studies of viral protein interactions with G3BPs to inhibit SGs have led to the speculation that plant viruses also evade host cellular processes by inhibiting SGs formation. The nuclear shuttle protein (NSP) of begomo ablution mosaic virus (AbMV), has a FVSF motif which interacts with the G3BP-like protein of Arabidopsis thaliana (AtG3BP) which is responsible for SGs assembly in plants during cellular stress and thereby inhibiting SGs. Moreover, the NSP of pea necrotic yellow dwarf virus (PNYDV) harbors a FNSGF motif which also interacts with AtG3BP [118]. These findings support the speculation that SGs assembly is conserved in plants and mammalian cells and plant viruses also inhibit SGs formation by interacting with G3BPs to invade host cellular responses in response to infection. The FGDF-mediated G3BP binding interactions described here represent an attractive target for therapeutic interventions against a range of diverse viral infections.
5.4. Viral targeting of G3BPs by proteases
It is interesting that unlike alphaviruses, flaviviruses and HCV, and the coronavirus systems discussed above, viruses like picornaviruses, cleave SG proteins, such as G3BPs, to inhibit SGs formation [98]. During poliovirus infection (PV), the viral 3C protease splits the NTF2-like domain and RRM domains of G3BPs by cleaving it after residue Q325, which subsequently inhibits SGs formation [119]. Encephalomyocarditis virus (EMCV) shares the same mechanism with PV to cleave G3BP1 after residue Q325 with specificity identical to that of the PV 3C protease. This was confirmed by experiments in which a mutation of this residue prevented cleavage of G3BP1 by EMCV [120]. G3BP1 associates directly to the three specific sequences of the internal ribosome entry site (IRES) elements of foot and mouth disease virus (FMDV) and the virus uses a similar mechanism to poliovirus to block G3BPs activity. During FMDV infection G3BP1 is also cleaved by its protease, 3C (3Cpro), yielding two fragments of G3BP (Ct-G3BP1, Nt-G3BP1) [121].
5.5. Viral inactivation of G3BPs by RNA
Viral RNA-host protein interactions are essential for the replication of RNA viruses like flaviviruses (a genus of positive strand RNA-viruses), which include the life threatening vector-borne human pathogens such as West Nile virus (WSNV) and Dengue virus (DENV). Recently it has been shown that all flaviviruses have non-coding subgenomic flaviviral RNA (sfRNA), formed by the incomplete degradation of viral 3′-UTR by cellular exonuclease XRN1 [122]. Studies on the Kunjin strain (KUNV) of WSNV showed that sfRNA antagonized IFN-mediated antiviral activity [122,123]. DENV-2 sfRNA binds to G3BP1, G3BP2 and Caprin1, impeding their role in innate immunity by inhibiting SGs which, in turn, inhibits translation of ISGs mRNA. G3BP1, G3BP2 and Caprin1 are novel regulators of antiviral responses as they are required for the efficient translation of interferon stimulated genes (ISGs) including protein kinase R (PKR) and interferon-induced transmembrane protein 2 (IFITM2) [34] and this role of G3BPs in ISGs mRNA translation is supported by the study reporting a role of G3BP1 and G3BP2 in the translational regulation of IFITM1-3 mRNAs. [66]. Furthermore, G3BP1 also restricts HIV-1 infection by interacting with HIV-1 RNA in the cytoplasm. This interaction sequesters the viral transcripts, thereby inhibiting protein synthesis or packaging [124], however to invade this response, HIV-1 also blocks SGs formation and dismantles the pre-formed SGs.
All these reports show that many viruses target and manipulate G3BPs to invade cellular responses that would inhibit the virus. Viruses do this by either targeting G3BPs with the result of disrupting SGs formation or by inhibiting the translation of the ISGs through interactions with G3BPs. Either way, disrupting G3BPs functions appears to be a common theme adopted by viruses to avoid cellular and host antiviral responses.
6. Conclusion
In our original review [1] the discovery of G3BPs had only just begun and like its namesake, Rasputin, we concluded that its activities were promiscuous and seemed to lack a consistent theme. More than a decade of research has failed to tame G3BPs but we now see a more focused role for the protein in cellular biology and it has now been demonstrated that G3BPs are involved in multiple cell signalling pathways. The most commonly reported mechanism of action for G3BPs is to interact with many cellular RNAs, controlling their fate in response to environmental and cellular stimuli. However, the binding elements to which G3BPs bind are yet to be clearly identified and there does not appear to be a clear consensus on this yet. Early evidence suggested that G3BPs might bind to target mRNA in the nucleus suggesting that it might be a shuttle protein and this is supported by its co-localization to transcriptional active sites. Is the role of G3BPs that of a chaperone to transport mRNA cargo to appropriate cytoplasmic structures including, but not exclusively, the SGs? The dual roles that G3BPs play in both the translation and degradation of transcripts still appears to be contradictory but does seem to be contextual based depending on cell type, protein partners and possibly RNA elements within its targets. The role of G3BPs as components of SGs is irrefutable, however, their role there still remains contentious. Intuitively, SGs are seen as a cellular response that allows the cell the opportunity to respond and recover to environmental stress. This is seen best by the fact that viruses need to disrupt G3BPs function to avoid a cell based anti-viral response. For certain, viruses hunt down G3BPs and use an impressive array of mechanisms to do so but how does G3BPs hunt down its targets? A hint to this might reside in its myriad of binding partners which appear to facilitate target specificity. The next decade will undoubtedly reveal G3BPs' secrets and with this will coincide with the exciting challenge of understanding how we can use G3BPs to fight diseases such as cancer and viral infection.
Conflict of interest
The authors declare that they have no conflict of interest with any company that could benefit from the findings of this manuscript.
Transparency document
Transparency document.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Irvine K., Stirling R., Hume D., Kennedy D. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 2004;48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 2.Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodsell D.S. The molecular perspective: the ras oncogene. Oncologist. 1999;4:263–264. [PubMed] [Google Scholar]
- 4.Kennedy D., French J., Guitard E., Ru K., Tocque B., Mattick J. Characterization of G3BPs: tissue specific expression, chromosomal localisation and rasGAP (120) binding studies. J. Cell. Biochem. 2001;84:173–187. doi: 10.1002/jcb.1277. [DOI] [PubMed] [Google Scholar]
- 5.Gallouzi I.-E., Parker F., Chebli K., Maurier F., Labourier E., Barlat I., Capony J.-P., Tocque B., Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol. Cell. Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annibaldi A., Dousse A., Martin S., Tazi J., Widmann C. Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317–326 sequence does not involve G3BP1. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suyama M., Doerks T., Braun I.C., Sattler M., Izaurralde E., Bork P. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 2000;1:53–58. doi: 10.1093/embo-reports/kvd009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai K., Oubridge C., Ito N., Avis J., Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem. Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 9.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Vognsen T., Moller I.R., Kristensen O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French J., Stirling R., Walsh M., Kennedy H.D. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem. J. 2002;34:223–231. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- 12.Barnes C.J., Li F., Mandal M., Yang Z., Sahin A.A., Kumar R. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res. 2002;62:1251–1255. [PubMed] [Google Scholar]
- 13.Prigent M., Barlat I., Langen H., Dargemont C. IκBα and IκBα/NF-κB complexes are retained in the cytoplasm through interaction with a novel partner, Ras-GAP SH3-binding protein 2. J. Biol. Chem. 2000;275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 14.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Macara I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booker G.W., Gout I., Kristina A., Driscoll P.C., Boyd J., Waterfield M.D., Campbell I.D. Solution structure and ligand-binding site of the SH3 domain of the p85α subunit of phosphatidylinositol 3-kinase. Cell. 1993;73:813–822. doi: 10.1016/0092-8674(93)90259-s. [DOI] [PubMed] [Google Scholar]
- 17.Kay B.K., Williamson M.P., Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 18.Clery A., Blatter M., Allain F.H.T. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 19.McBride A.E., Silver P.A. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 20.Shen E.C., Henry M.F., Weiss V.H., Valentini S.R., Silver P.A., Lee M.S. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols R.C., Wang X.W., Tang J., Hamilton B.J., High F.A., Herschman H.R., Rigby W.F.C. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 2000;256:522–532. doi: 10.1006/excr.2000.4827. [DOI] [PubMed] [Google Scholar]
- 22.Friend L.R., Landsberg M.J., Nouwens A.S., Wei Y., Rothnagel J.A., Smith R. Arginine methylation of hnRNP A2 does not directly govern its subcellular localization. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikkavilli R.K., Malbon C.C. Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J. Cell Sci. 2011;124:2310–2320. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bikkavilli R.K., Malbon C.C. Wnt3a-stimulated LRP6 phosphorylation is dependent upon arginine methylation of G3BP2. J. Cell Sci. 2012;125:2446–2456. doi: 10.1242/jcs.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai W.-C., Gayatri S., Reineke L.C., Sbardella G., Bedford M.T., Lloyd R.E. Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai W.-C., Reineke L.C., Jain A., Jung S.Y., Lloyd R.E. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule–nucleating protein G3BP1. J. Biol. Chem. 2017;292:18886–18896. doi: 10.1074/jbc.M117.800706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandran B., Stabley J.N., Cheng S.-L., Behrmann A.S., Gay A., Li L., Mead M., Kozlitina J., Lemoff A., Mirzaei H., Chen Z., Towler D.A. A GTPase-activating protein binding protein (G3BP1)/antiviral protein relay conveys arteriosclerotic Wnt signals in aortic smooth muscle cells. J. Biol. Chem. 2018;293:7942–7968. doi: 10.1074/jbc.RA118.002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa M., Ochem A., Staub A., Falaschi A. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 1999;27:817–821. doi: 10.1093/nar/27.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tourriere H., Gallouzi I.-E., Chebli K., Capony J.P., Mouaikel J., van der Geer P., Tazi J. RasGAP-associated endoribonuclease G3BP: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guitard E., Parker F., Millon R., Abecassis J., Tocqué B. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett. 2001;162:213–221. doi: 10.1016/s0304-3835(00)00638-8. [DOI] [PubMed] [Google Scholar]
- 31.Kociok N., Esser P., Unfried K., Parker F., Schraermeyer U., Grisanti S., Toque B., Heimann K. Upregulation of the RAS-GTPase activating protein (GAP)-binding protein (G3BP) in proliferating RPE cells. J. Cell. Biochem. 1999;74:194–201. [PubMed] [Google Scholar]
- 32.Zekri L., Chebli K., Tourriere H., Nielsen F.C., Hansen T.V., Rami A., Tazi J. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol. Cell. Biol. 2005;25:8703–8719. doi: 10.1128/MCB.25.19.8703-8716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winslow Sofia, Leandersson K., Larsson C. Regulation of PMP22 mRNA by G3BP1 affects cell proliferation in breast cancer cells. Mol. Cancer Res. 2013;12:156. doi: 10.1186/1476-4598-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidet K., Dadlani D., Garcia-Blanco M.A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei S.C., Fattet L., Tsai J.H., Guo Y., Pai V.H., Majeski H.E., Chen A.C., Sah R.L., Taylor S.S., Engler A.J., Yang J. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., Ma Y., Zhang S., Liu H., He H., Li N., Gong Y., Zhao S., Jiang J.-d., Shao R.-g. Involvement of Ras GTPase-activating protein SH3 domain-binding protein 1 in the epithelial-to-mesenchymal transition-induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget. 2015;6:17039–17053. doi: 10.18632/oncotarget.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniuchi K., Nishimori I., Hollingsworth M.A. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol. Cancer Res. 2011;9:856–866. doi: 10.1158/1541-7786.MCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 38.Soncini Chiara, Berdo I., Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;22:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 39.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgescu M.-M. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Yanping, Wernyj R.P., Norton Darrell D., Precht Patricia, Seminario Maria-Cristina, Wange Ronald L. Modulation of specific protein expression levels by PTEN: identification of AKAP121, DHFR, G3BP, Rap1, and RCC1 as potential targets of PTEN. Oncogene. 2005;24:3819–3829. doi: 10.1038/sj.onc.1208527. [DOI] [PubMed] [Google Scholar]
- 42.Kim M.M., Wiederschain D., Kennedy D., Hansen E., Yuan Z.M. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP) Oncogene. 2007;26:4209–4215. doi: 10.1038/sj.onc.1210212. [DOI] [PubMed] [Google Scholar]
- 43.Mao C., Wang X., Liu Y., Wang M., Yan B., Jiang Y., Shi Y., Shen Y., Liu X., Liai W., Yang R., Xiao D., Cheng Y., Liu S., Zhou H., Cao Y., Yu W., Muegge K., Yu H., Tao Y. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–3496. doi: 10.1158/0008-5472.CAN-17-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashikari D., Takayama K., Obinata D., Urano T., Inoue S., Takahashi S. 276 A novel androgen-responsive gene, G3BP2, induces therapeutic-resistance in castration-resistant prostate cancer cells. Eur. Urol. Suppl. 2015;14 [Google Scholar]
- 45.Ashikari D., Takayama K., Tanaka T., Suzuki Y., Obinata D., Fujimura T., Urano T., Takahashi S., Inoue S. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer. Oncogene. 2017;36:6272. doi: 10.1038/onc.2017.225. [DOI] [PubMed] [Google Scholar]
- 46.Takayama K.-I., Suzuki T., Tanaka T., Fujimura T., Takahashi S., Urano T., Ikeda K., Inoue S. TRIM25 enhances cell growth and cell survival by modulating p53 signals via interaction with G3BP2 in prostate cancer. Oncogene. 2018;37:2165–2180. doi: 10.1038/s41388-017-0095-x. [DOI] [PubMed] [Google Scholar]
- 47.Takayama K.-I., Suzuki T., Fujimura T., Takahashi S., Inoue S. Association of USP10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol. Cancer Res. 2018;16:846–856. doi: 10.1158/1541-7786.MCR-17-0471. [DOI] [PubMed] [Google Scholar]
- 48.Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 49.Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Zhang S., He H., Zhang C., Yu D., Shao R. Downregulation of G3BPs inhibits the growth, migration and invasion of human lung carcinoma H1299 cells by suppressing the Src/FAK-associated signaling pathway. Cancer Gene Ther. 2013;20:622–629. doi: 10.1038/cgt.2013.62. [DOI] [PubMed] [Google Scholar]
- 51.Taniuchi K., Nishimori I., Hollingsworth M.A. Intracellular CD24 inhibits cell invasion by posttranscriptional regulation of BART through interaction with G3BP. Cancer Res. 2011;71:895–905. doi: 10.1158/0008-5472.CAN-10-2743. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H.-Z., Liu J.-G., Wei Y.-P., Wu C., Cao Y.-K., Wang M. Expression of G3BP and RhoC in esophageal squamous carcinoma and their effect on prognosis. World J. Gastroenterol. 2007;13:4126–4130. doi: 10.3748/wjg.v13.i30.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Somasekharan S.P., El-Naggar A., Leprivier G., Cheng H., Hajee S., Grunewald T.G.P., Zhang F., Ng T., Delattre O., Evdokimova V., Wang Y., Gleave M., Sorensen P.H. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beheshtizadeh M., Moslemi E. Analysis of G3BP1 and VEZT expression in gastric cancer and their possible correlation with tumor clinicopathological factors. J. Gastric Cancer. 2017;17:43–51. doi: 10.5230/jgc.2017.17.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dou N., Chen J., Yu S., Gao Y., Li Y. G3BP1 contributes to tumor metastasis via upregulation of Slug expression in hepatocellular carcinoma. Am. J. Cancer Res. 2016;6:2641–2650. [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta N., Badeaux M., Liu Y., Naxerova K., Sgroi D., Munn L.L., Jain R.K., Garkavtsev I. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc. Natl. Acad. Sci. 2017;114:1033–1038. doi: 10.1073/pnas.1525387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ocana Oscar H., Córcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., Barrallo-Gimeno A., Cano A., Nieto M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H., Zhang S., He H., Zhao W., Chen J., Shao R.-g. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012;103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H., Zhang S., He H., Zhang C., Chen Y., Yu D., Chen J., Shao R. RasGAP-derived peptide GAP159 enhances cisplatin-induced cytotoxicity and apoptosis in HCT116 cells. Acta Pharm. Sin. B. 2014;4:128–134. doi: 10.1016/j.apsb.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shim J.-H., Su Z.-Y., Chae J.-I., Kim D.J., Zhu F., Ma W.-Y., Bode A.M., Yang C.S., Dong Z. Epigallocatechin gallate suppresses lung cancer cell growth through Ras–GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev. Res. 2010;3:670–679. doi: 10.1158/1940-6207.CAPR-09-0185. [DOI] [PubMed] [Google Scholar]
- 61.Oi N., Yuan J., Malakhova M., Luo K., Li Y., Ryu J., Zhang L., Bode A.M., Xu Z., Li Y., Lou Z., Dong Z. Resveratrol induces apoptosis by directly targeting Ras-GTPase activating protein SH3 domain binding protein 1 (G3BP1) Oncogene. 2015;34:2660–2671. doi: 10.1038/onc.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atlas R., Behar L., Elliott E., Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 2004;89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 63.Lypowy J., Chen I.-Y., Abdellatif M. An alliance between Ras GTPase-activating protein, filamin C, and Ras GTPase-activating protein SH3 domain-binding protein regulates myocyte growth. J. Biol. Chem. 2005;280:25717–25728. doi: 10.1074/jbc.M414266200. [DOI] [PubMed] [Google Scholar]
- 64.Ortega A.D., Willers I.M., Sala S., Cuezva J.M. Human G3BP1 interacts with β-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 2010;123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- 65.Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., Schrader J.W. Distinct structural features of Caprin-1 mediate its interaction with G3BP1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Umber A., Kennedy D. 2018. G3BP1 and G3BP2 Regulate Translation of Interferon Stimulated Genes; IFITM1, IFITM2 and IFITM3 in Cancer Cell Line MCF7. (submitted) [DOI] [PubMed] [Google Scholar]
- 67.He M., Yang Z., Abdellatif M., Sayed D. GTPase activating protein (SH3 domain) binding protein 1 regulates the processing of microRNA-1 during cardiac hypertrophy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong H.-Q., Lu J., Fang X.-L., Zhang Y.-H., Cai Y., Yuan J., Liu P.-Q., Ye J.-T. G3BP2 is involved in isoproterenol-induced cardiac hypertrophy through activating the NF-κB signaling pathway. Acta Pharmacol. Sin. 2017;39:184–194. doi: 10.1038/aps.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwok H.-H., Poon P.-Y., Mak K.H.-M., Zhang L.-Y., Liu P., Zhang H., Mak N.-K., Yue P.Y.-K., Wong R.N.-S. Role of G3BP1 in glucocorticoid receptor-mediated microRNA-15b and microRNA-23a biogenesis in endothelial cells. Cell. Mol. Life Sci. 2017;74:3613–3630. doi: 10.1007/s00018-017-2540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin S., Bellora N., González-Vallinas J., Irimia M., Chebli K., de Toledo M., Raabe M., Eyras E., Urlaub H., Blencowe B.J., Tazi J. Preferential binding of a stable G3BP ribonucleoprotein complex to intron-retaining transcripts in mouse brain and modulation of their expression in the cerebellum. J. Neurochem. 2016;139:349–368. doi: 10.1111/jnc.13768. [DOI] [PubMed] [Google Scholar]
- 71.Zhao B., Li H., Liu J., Han P., Zhang C., Bai H., Yuan X., Wang X., Li L., Ma H., Jin X., Chu Y. MicroRNA-23b targets Ras GTPase-activating protein SH3 domain-binding protein 2 to alleviate fibrosis and albuminuria in aiabetic nephropathy. J. Am. Soc. Nephrol. 2016 Sep;27(9):2597–2608. doi: 10.1681/ASN.2015030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Angenstein F., Evans A.M., Settlage R.E., Moran S.T., Ling S.C., Klintsova A.Y., Shabanowitz J., Hunt D.F., Greenough W.T. A receptor for activated C kinase is part of messenger ribonucleoprotein complexes associated with polyA-mRNAs in neurons. J. Neurosci. 2002;22:8827–8837. doi: 10.1523/JNEUROSCI.22-20-08827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson P., Kedersha N., Ivanov P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta, Gene Regul. Mech. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and out of translation. Mol. Cell. 2009;36:932. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 77.Kim W.J., Back S.H., Kim V., Ryu I., Jang S.K. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W., Simarro M., Kedersha N., Anderson P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol. Cell. Biol. 2004;24:10718–10732. doi: 10.1128/MCB.24.24.10718-10732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 80.McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.-J., Anderson P., Kaufman R.J. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 81.Donnelly N., Gorman A.M., Gupta S., Samali A. The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 83.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))–deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas M.G., Loschi M., Desbats M.A., Boccaccio G.L. RNA granules: the good, the bad and the ugly. Cell. Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., Anderson P. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aguilera-Gomez A., Zacharogianni M., van Oorschot M.M., Genau H., Grond R., Veenendaal T., Sinsimer K.S., Gavis E.R., Behrends C., Rabouille C. Phospho-Rasputin stabilization by Sec16 is required for stress granule formation upon amino acid starvation. Cell Rep. 2017;20:935–948. doi: 10.1016/j.celrep.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mazroui R., Sukarieh R., Bordeleau M.-E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 90.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 92.Reineke L.C., Dougherty J.D., Pierre P., Lloyd R.E. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 2012;23:3499–3510. doi: 10.1091/mbc.E12-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vanderweyde T., Yu H., Varnum M., Liu-Yesucevitz L., Citro A., Ikezu T., Duff K., Wolozin B. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. 2012;32:8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 95.Aulas A., Caron G., Gkogkas C.G., Mohamed N.-V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J.A., Vande Velde C. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 2015;209:73–84. doi: 10.1083/jcb.201408092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kobayashi T., Winslow S., Sunesson L., Hellman U., Larsson C. PKCα binds G3BP2 and regulates stress granule formation following cellular stress. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao X., Ge L., Shao J., Su C., Zhao H., Saarikettu J., Yao X., Yao Z., Silvennoinen O., Yang J. Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett. 2010;584:3525–3532. doi: 10.1016/j.febslet.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lloyd R.E. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip. Rev. RNA. 2013;4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reineke L.C., Lloyd R.E. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim D.Y., Reynaud J.M., Rasalouskaya A., Akhrymuk I., Mobley J.A., Frolov I., Frolova E.I. New world and old world alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katsafanas G.C., Moss B. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- 102.Yi Z., Pan T., Wu X., Song W., Wang S., Xu Y., Rice C.M., MacDonald M.R., Yuan Z. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J. Virol. 2011;85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yi Z., Fang C., Pan T., Wang J., Yang P., Yuan Z. Subproteomic study of hepatitis C virus replicon reveals Ras-GTPase-activating protein binding protein 1 as potential HCV RC component. Biochem. Biophys. Res. Commun. 2006;350:174–178. doi: 10.1016/j.bbrc.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 104.Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 2006;347:683–691. doi: 10.1016/j.bbrc.2006.06.144. [DOI] [PubMed] [Google Scholar]
- 105.Garaigorta U., Heim M.H., Boyd B., Wieland S., Chisari F.V. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012;86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Panas M.D., Varjak M., Lulla A., Er Eng K., Merits A., Karlsson Hedestam G.B., McInerney G.M. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 2012;23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Panas M.D., Ahola T., McInerney G.M. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J. Virol. 2014;88:5888–5893. doi: 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gorchakov R., Garmashova N., Frolova E., Frolov I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008;82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fros J.J., Domeradzka N.E., Baggen J., Geertsema C., Flipse J., Vlak J.M., Pijlman G.P. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J. Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cristea I.M., Carroll J.-W.N., Rout M.P., Rice C.M., Chait B.T., MacDonald M.R. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 2006;281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 111.Frolova E., Gorchakov R., Garmashova N., Atasheva S., Vergara L.A., Frolov I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cristea I.M., Rozjabek H., Molloy K.R., Karki S., White L.L., Rice C.M., Rout M.P., Chait B.T., MacDonald M.R. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 2010;84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scholte F.E.M., Tas A., Albulescu I.C., Zusinaite E., Merits A., Snijder E.J., van Hemert M.J. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J. Virol. 2015;89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fros J.J., Geertsema C., Zouache K., Baggen J., Domeradzka N., van Leeuwen D.M., Flipse J., Vlak J.M., Failloux A.-B., Pijlman G.P. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasit. Vectors. 2015;8:464. doi: 10.1186/s13071-015-1070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panas M.D., Schulte T., Thaa B., Sandalova T., Kedersha N., Achour A., McInerney G.M. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Valiente-Echeverria F., Melnychuk L., Vyboh K., Ajamian L., Gallouzi I.E., Bernard N., Mouland A.J. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat. Commun. 2014;5 doi: 10.1038/ncomms5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matthews J.D., Frey T.K. Analysis of subcellular G3BP redistribution during rubella virus infection. J. Gen. Virol. 2012;93:267–274. doi: 10.1099/vir.0.036780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krapp S., Greiner E., Amin B., Sonnewald U., Krenz B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res. 2017;227:6–14. doi: 10.1016/j.virusres.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 119.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 120.Ng C.S., Jogi M., Yoo J.-S., Onomoto K., Koike S., Iwasaki T., Yoneyama M., Kato H., Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J. Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Galan A., Lozano G., Pineiro D., Martinez-Salas E. G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. FEBS J. 2017;284:3202–3217. doi: 10.1111/febs.14184. [DOI] [PubMed] [Google Scholar]
- 122.Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 123.Schuessler A., Funk A., Lazear H.M., Cooper D.A., Torres S., Daffis S., Jha B.K., Kumagai Y., Takeuchi O., Hertzog P., Silverman R., Akira S., Barton D.J., Diamond M.S., Khromykh A.A. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cobos Jimenez V., Martinez F.O., Booiman T., van Dort K.A., van de Klundert M.A.A., Gordon S., Geijtenbeek T.B.H., Kootstra N.A. G3BP1 restricts HIV-1 replication in macrophages and T-cells by sequestering viral RNA. J. Virol. 2015;486:94–104. doi: 10.1016/j.virol.2015.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.