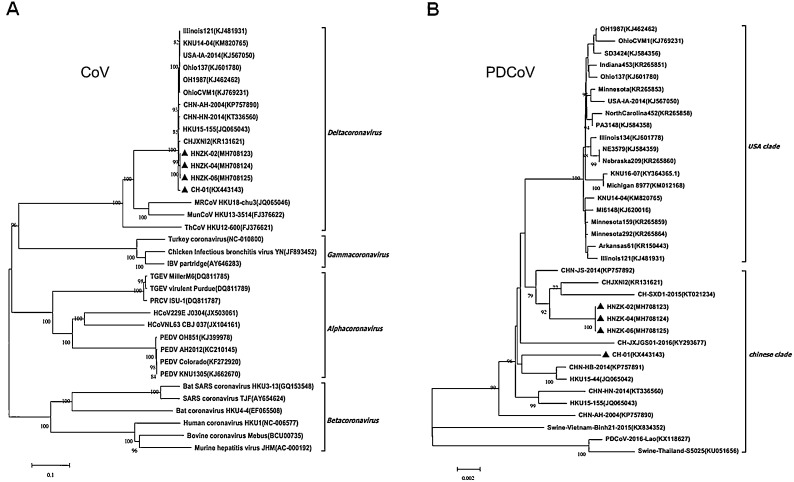
Fig. 3.
Phylogenetic analysis based on the complete genome sequences of PDCoV strains. (A) The nucleotide sequences of the full-length genomes from four coronavirus genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus). (B). Phylogenetic analyses of complete nucleotide sequences of 4 PDCoVs from this study and some published PDCoV sequences in Genbank. The phylogenetic tree was constructed from the aligned nucleotide sequences using the neighbour-joining method with MEGA 6.06 software (http://www.megasoftware.net). Bootstrap values were calculated with 1000 replicates. Reference sequences obtained from GenBank are indicated by strain names and GenBank accession numbers. The PDCoV isolates identified in this study are indicated with black triangles. Scale bars indicate nucleotide substitutions per site.