Abstract
Positive controls are essential for PCR reliability and are challenging to obtain for rare, exotic and/or emerging pathogens and pose biosafety risks if manufactured using infectious pathogens. Custom synthetic DNA inserts can be designed de novo in tandems of forward and reverse complement priming sequences to be inserted in circular plasmid vectors. To test this concept, artificial positive controls (APCs) for use in PCR were synthesized to contain primer sequences targeting four viruses (Barley yellow dwarf virus, Soilborne wheat mosaic virus, Triticum mosaic virus and Wheat streak mosaic virus) pathogenic to wheat and, as internal control, the plant mitochondrial nad5 gene. Thermodynamics and folding parameters of twenty-four APC inserts were assessed in silico. Two thermodynamically different APCs, designated optimal and sub-optimal, were cloned and tested using end point PCR. The optimal APC had a 100% amplification rate, while only 92% of virus-infected plant tissues, commonly used as reference positive controls, amplified. An array of APC priming sequences from different organisms and/or previously tested primers can be accommodated in a large and flexible number of positive control targets. APCs will streamline and standardize routine PCR, improve reliability and biosafety, and create opportunities for development and commercialization of new synthetic positive control sequences.
Keywords: Synthetic DNA, Nucleic acids thermodynamics, Detection, Diagnostics
Highlights
-
•
Controls are essential for PCR reliability in diagnostics and microbial forensics.
-
•
Controls are challenging to obtain for rare, exotic and/or emerging pathogens.
-
•
Positive controls pose biosafety risks if manufactured using infectious pathogens.
-
•
Tandems of forward and reverse primers were engineered as synthetic DNA inserts.
-
•
Synthetic controls were demonstrated in silico and subsequently in vitro by PCR.
1. Introduction
The advent of synthetic biology offers new possibilities for designing, engineering and developing artificial molecules to reproduce natural biological processes, and for creating interchangeable biological parts to assemble new devices and systems (Louw et al., 2011, Schmith, 2008, Stähler et al., 2006). The creation of synthetic artificial controls for a variety of microbial detection and disease diagnostic assays is a new alternative to the use of in vivo positive controls. Plasmid construct-based synthetic controls harboring a custom and de novo designed insert can enhance technologies such as polymerase chain reaction (PCR) to improve biosafety, speed of processing and overall detection without compromising sensitivity and specificity.
Examples of artificial plasmid constructs containing partial or complete sequences of a pathogen (or multiple pathogens) of interest are already in use or reported. Charrel et al. (2004) designed and constructed plasmids containing specific sequences of category A agents of bioterrorism, including exogenic sequences with inserted NotI sites and primers designed to amplify agent specific sequences and probes to detect specifically each sequence. Steps toward construction included: PCR amplification of targets and incubation to add restriction sites; column purification to discard residual small DNA fragments remnant from the restriction reaction; overnight incubation in the presence of T4 DNA ligase at 4 °C; and cloning of the final product into a plasmid. Similarly, Inoue et al. (2004) constructed a recombinant plasmid harboring fragments of the pag and cap genes of Bacillus anthracis distinguishable from the authentic sequences by the presence of restriction enzyme sites, the EcoRV for the pag gene and the BamHI for the cap gene, respectively. Siegling et al. (1994) reported the construction of a multispecific control fragment containing 14 different templates for use in competitive PCR quantification of rat gene expression. Primer pairs did not exhibit 3′-complementarity and spanned over one or more introns to facilitate distinguishing the amplicons obtained from cDNA from those from genomic DNA. The construction was made using complementary 40mer oligonucleotides cloned in a plasmid containing binding sites for both sense primers. Chomič et al. (2010) reported the construction of synthetic positive controls for the detection of Luteoviridae through RT-PCR. The sequences of forward and reverse primers were arranged into two fragments, and inserted in a plasmid. The two fragments were 152 and 161 bp long; however, the synthesis of oligonucleotides greater than 100 bases was not possible. Therefore, each fragment was synthesized as two separate oligonucleotides overlapping by 20 bp. Smith et al. (2006) also described a labor intensive procedure to produce synthetic controls including probes for only two targets, SARS coronavirus and rodent glyceraldehyde-3-phosphate dehydrogenase (rGAPDH), for realtime RT-PCR (TaqMan). The method used the bacteriophage T7, RNA polymerase binding site included at the 5′ terminus of the oligonucleotide to allow RNA transcription of the oligonucleotide. The reported strategies in all of these examples provide a safe and reproducible positive-control DNA template and also allow the detection of possible cross contamination, but their development in vitro was laborious and the constructs have been limited to a small number of targets. The approach to be described here is unique in its combinatorial primer design, enlarged capacity for targets, flexibility to rearrange and update, and elimination of pathogen sequences (intron, overlapping, etc.,) except for the primer sequences.
Nucleic acid-based molecular assays such PCR have become preferred diagnostic platforms, particularly due to recent enhancements (Alvarez, 2004, Henson and French, 1993). Although PCR has been adapted for routine testing for pathogens of interest (Miller et al., 2009), positive controls may be challenging to obtain (Smith et al., 2006) because not all proteins or pathogens are available as positive controls. Moreover, positive controls are especially difficult to obtain for exotic emerging pathogens, which are not commercially available in most cases, and require time-consuming obtaining of permits and clearances (Nechvatal et al., 2008, Smith et al., 2006). Infected tissue, purified or cultured pathogen, and/or purified or expressed proteins are positive controls commonly used. However, well-documented risks associated with the handling or shipping of infected sample controls, particularly those carrying highly pathogenic microorganisms that are efficiently transmitted (Charrel et al., 2004, Inoue et al., 2004, Siegling et al., 1994, Smith et al., 2006), threaten the biosafety of their use.
The aim of this research is to assess the suitability of a relatively new concept of customized synthetic DNA inserts consisting of linear arrays of primer sequences designed from a variety of genomes or targets of value for detection, diagnostics and research.
2. Materials and methods
2.1. Primer sequences
Sequences of five primer sets were selected to design and synthesize APC inserts. The five primer sets target four wheat viruses, Barley yellow dwarf virus (BYDV), Soilborne wheat mosaic virus (SBWMV), Triticum mosaic virus (TriMV), and Wheat streak mosaic virus (WSMV) and the putative plant internal control mitochondrial nad5 gene. Primers for BYDV and SBWMV were designed in this study. The Genbank accession numbers incorporated into consensus sequences for primer design of BYDV, SBWMV (RNA2), TriMV (Data not shown) and WSMV (Data not shown) are presented in Supplementary Table 2. Primer sequences targeting nad5 are from previously reported literature (Menzel et al., 2002). All consensus and primer design regions were obtained after complete alignment of multiple nucleotide sequences using ClustalX (Larkin et al., 2007). Forward and reverse primers were designed using Primer3 (Rozen and Skaletsky, 2000) following reported parameters (Arif and Ochoa-Corona, 2012). The ΔG, secondary structures, self- and heterodimer formation of each set of primers were examined after matching and analyzing the outputs from the Primer3 ‘any’ self-complementarity score, which was taken as a measure of its tendency to anneal to itself or form secondary structure, and mFold (Rozen and Skaletsky, 2000, Zuker, 2003). The specificity of each primer sequence was confirmed in silico by searching the NCBI nucleotide database using BLASTn (Altschul et al., 1990). The preliminary in vitro performance of all primer sets was tested by reverse transcription-PCR (RT-PCR) using virus infected plant tissue as positive controls for each targeted virus. Primers used for engineering APC inserts and performing PCR and RT-PCR are listed in Table 1 .
Table 1.
Primers used for designing the APC synthetic inserts, performing PCR of the targets within APCs, and RT-PCR of in vivo reference positive controlsa.
| Target | Primer ID | 5′-3′ sequence | Size of target amplicons (bp) |
Reference | ||
|---|---|---|---|---|---|---|
| Reference positive controla | Optimal APC | Sub-optimal APC | ||||
| Barley yellow dwarf virus | BYDV-F1 | CAACACCGGAACCGAAAC | 128 | 121 | 123 | This study |
| BYDV-R1 | TGGAGTGCCGATATACTCAAA | |||||
| Soil-borne wheat mosaic virus | SBWMV-F1 | ATCATTGTCCCGCTGTTCTC | 92 | 120 | 122 | This study |
| SBWMV-R1 | GCACACCTGCTTCTTTCCA | |||||
| Wheat streak mosaic virus | WSMV-F2 | GAACTCAAAGCACCCACAA | 218 | 122 | 122 | Arif et al. (unpublished) |
| WSMV-R2 | ACCACCACATCAACCTCCTC | |||||
| Triticum mosaic virus | TriMV-F1 | AACACAACGCACGACTTTCT | 198 | 122 | 122 | Arif et al. (unpublished) |
| TriMV-R1 | GCAACCCATTCCTTCTTCC | |||||
| NADH dehydrogenase subunit 5 | Nad5-s | GATGCTTCTTGGGGCTTCTTGTT | 181 | 124 | 124 | Menzel et al. (2002) |
| Nad5-as | CTCCAGTCACCAACATTGGCATAA | |||||
Infected plant tissue.
2.2. The APC concept and in silico assessment of APC inserts
The APC concept consists of a DNA insert that can be custom designed de novo and synthesized artificially. APC inserts contain primer sequences in series, an upstream component of 5′-3′ forward primers followed by a second (downstream) of 5′-3′ reverse complement primer sequences. The APC insert is ligated to a selected restriction enzyme site within a multiple cloning site of a chosen plasmid vector for circularization, yielding a functionally clonable APC construct (Fig. 1 ). The APC approach also allows the inclusion of a large number of targets arranged in groups or tandems of APC inserts (Fig. 1-B).
Fig. 1.
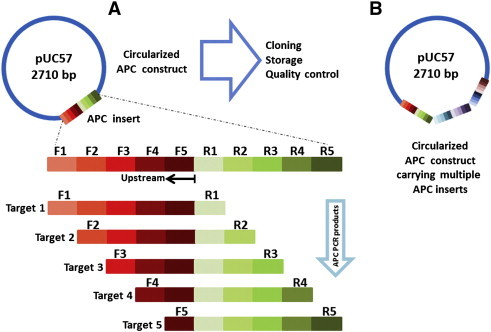
The APC concept. (A) The APC insert is made by custom synthesized tandems of forward and reverse complement priming sequences ligated into the multiple cloning site of pUC57 to make a circular and clonable APC construct. Each PCR product amplified from the APC construct has a unique identifiable sequence useful for further PCR product verification and quality control. (B) An APC construct carrying multiple custom synthetic APC inserts. Not to scale.
APC inserts were designed combining a tandem of forward and reverse complement of primer sequences in different sequence orders for the nad5 internal control and the four targeted viruses (Table 2 ). Out of the 120 possible combinations, 24 with nad5 at the 5′ position were selected. Each combination was 203 bp long and all theoretically able to generate PCR products from 120 to 124 bp (Table 1 — size of target amplicons). All primer sequence combinations within the APC insert were determined using a SAS 9.2 permutation code in SAS 9.2 (SAS Institute Inc., Cary, NC). Because forward and reverse complement primer sequences were arranged in combinatorial tandems, the expected product of each primer set contains the amplifying primer sequences and other primer sequences within the APC insert, giving to each APC product a specific sequence identity useful for monitoring quality control and reaction specificity (Fig. 1-A).
Table 2.
Sequences of the optimal (A) and sub-optimal (B) APCs showing primer sequence organization. Upstream forward primer sequences are bold, followed by the complement reverse primer sequences downstream. Individual primer sequences are indicated by alternating underlines. Primer sets sequence order is, Optimal APC POS-2_7: Nad5, SBWMV, BYDV, TriMV, and WSMV, and suboptimal APC POS-2_1: Nad5, BYDV, SBWMV, TriMV, and WSMV. Restriction site (EcoRV) overhangs at the 5′ and 3′ termini of the APC inserts are not show.
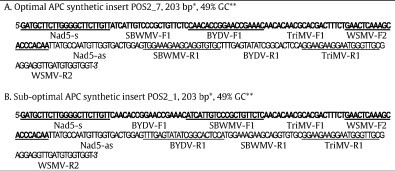
*Length and **GC content were calculated using Oligo Calculator (Dana–Farber Cancer Institute).
The twenty-four APC inserts were assessed in silico. Plot ΔG, and optimal free energy of predicted secondary structures were determined using mFOLD at 60 °C. The Plot ΔG or energy dot plot contains the superposition of all possible DNA folding and gives an overall visual of DNA folding facilitating the analysis.
2.3. Construction of APCs and assessment by PCR
Two APC sequences from the pool of 24 theoretical APC inserts were categorized as suboptimal (SubOpt) and optimal (Opt) and were cloned into plasmid vector pUC57. The criteria for selecting the two inserts were: ∆G far from 0 kcal mol− 1 and the presence of several secondary structures for the SubOpt insert, while the Opt APC insert had a ∆G value close to 0 kcal mol− 1 and few or no secondary structures. The two selected SubOpt and Opt APC inserts (POS-2_1 and POS-2_7 in Table 1 respectively) were synthesized and inserted at the EcoRV site of plasmid vector pUC57 and subsequently cloned in Escherichia coli (DH5α) by Genscript USA Inc. (Piscataway, NJ). The APC construct plasmids were serially diluted from 10 to 0.1 ng μL− 1 and assessed by end point PCR. PCR reactions were carried out in 20 μL volumes using GoTaq® Green Master Mix (Promega, Madison, WI) following the manufacturer's protocol. PCR cycling conditions were the same for all five primer sets in each of the two plasmids and consisted of an initial denaturation step at 95 °C for 3 min; followed by 40 cycles of 95 °C for 20 s, 56 °C for 20 s, and 72 °C for 20 s; and a final extension 72 °C for 5 min.
2.4. Reference plant positive controls
Reference plant positive controls consisted of total RNA extracts from 130 plant samples naturally infected with wheat viruses in a number of known combinations. The reference plant positive controls were provided by the Oklahoma State University (OSU) Plant Disease and Insect Diagnostic Laboratory (PDIDL) and were collected at the OSU Agronomy Research Station experimental plots, Stillwater, Oklahoma. Total RNA extractions were made using Trizol® Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's procedure. Infected plant RNAs were confirmed positive by RT-PCR using primers listed in Table 1. PCR products were electrophoresed in a 1.5% agarose gel in 1X TAE buffer, and amplicon sizes were estimated using a 1 kb plus ladder (Invitrogen, Carlsbad, CA).
2.5. Comparison of APC constructs and reference plant positive controls
The dataset consisted of 90 plant samples that tested RT-PCR positive to BYDV, four to SBWMV, two to TriMV, and 45 to WSMV within the 130 field samples, and 130 E. coli colonies containing the Opt APC. The Opt APC was enriched in E. coli (DH5α) and maintained and stored at − 80 °C for further assessment and statistical analysis, PCR amplification using APC constructs was compared to that of reference positive controls (infected plant tissue) by a Fisher's exact Test at 5% significance. Statistical analysis was performed using SAS 9.2.
2.6. APC compatibility with the use of a paper based DNA sampling matrix
The central element of an apparatus for biologic sample rapid collection, recovery, and convenient storage, also named elution-independent collection device (EICD) was used to assess the performance in PCR of APCs infiltrated into a soluble paper sampling matrix. EICD was developed for rapid (3 min.) sample collection and preparation of microorganisms for PCR, nucleic acid recovery and long-term room temperature storage (Josue-Caasi, 2012, Ochoa-Corona, 2012). Small disks (1.2 mm diameter) of sample from an EICD central element containing Opt APC at 0.1 ng uL− 1 were dissolved directly in liquid PCR reagents.
3. Results
3.1. Thermodynamics and folding of APC inserts
All designed APC inserts carried the same primer sequences but the order of these primers varied, leading to differences in thermodynamics and in the formation of predicted secondary structures. The plot ΔG values of the 24 APC synthetic inserts ranged from 0.2 to 1 kcal mol− 1, and the optimal energies of the secondary structure ranged from − 0.6 to 1.0 kcal mol− 1. All APC inserts have one, two or three predicted folding regions. The predicted secondary structure and ∆G value of the Opt and Sub Opt APCs are shown in the Supplementary Table 1. The Opt APC has a ∆G of 0.2 kcal mol− 1, and one possible secondary structure with optimal energy of − 0.6 kcal mol− 1 (Supplementary Fig. 1A). The SubOpt APC has a ∆G of 1.0 kcal mol− 1, and three possible secondary structures with optimal energies of 1.01, 1.21 and 1.31 kcal mol− 1 (Supplementary Fig. 1B, C and D). The synthetic inserts of both Opt and SubOpt APCs were 203 bp long with 49% GC as predicted and their sequences and primer organization are shown in Table 2. The expected size of PCR products from APC inserts ranged from 120 to 124 bp as predicted (Table 1, Fig. 2 ).
Supplementary Fig. 1.
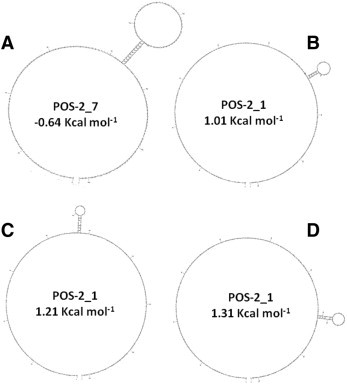
(A) Predicted DNA folding of the optimal APC synthetic insert POS-2_7 and free energy of major secondary structure. (B, C and D). Predicted DNA folding of sub-optimal APC synthetic insert POS-2_1. The three predicted secondary structures and their free energy is shown. All calculations were made by mFOLD at 60 °C, 10 mM Na+ and 1.5 mM Mg++.
Fig. 2.
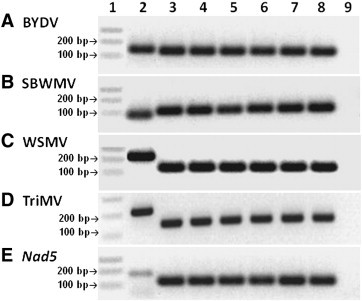
PCR amplification from infected plant reference positive controls, optimal APC POS-2_7 and sub-optimal APC POS-2_1. Gels, top to bottom, are: (A) BYDV, (B) SBWMV, (C) WSMV, (D) TriMV, and (E) Nad5. Lane 1 is a 1-Kb plus DNA ladder (Invitrogen, Carlsbad, CA). Lane 2 contains product obtained from cDNA from infected plant reference positive controls. Lanes 3, 4 and 5 are from optimal APC (POS-2_7) at 10, 1 and 0.1 ng μL− 1 respectively. Lanes 6, 7 and 8 are from suboptimal APC (POS-2_1) at 10, 1 and 0.1 ng μL− 1 respectively. Lane 9 is a blank negative control.
3.2. Efficacy of APC as a positive control
During a preliminary screening, all infected plant positive controls were RT-PCR positive for their respective viruses, BYDV, SBWMV, TriMV or WSMV when tested with the appropriate primers as well as with the primers for the plant internal control mitochondrial nad5 gene (Fig. 2, lane 2). Also, all targets of both optimal and sub-optimal APCs were amplified, with no detectable difference in the intensity of the amplicons produced by the two APCs regardless of plasmid DNA concentration (10 to 0.1 ng μL− 1) (Fig. 2, Lanes 3–8). As expected, differences in APC product sizes were observed among four of the five reference positive controls due to differences in size of the expected products and different distances between priming sequences within the APC synthetic inserts (Fig. 2). No positive matches were found in the GenBank database after alignment using BLASTn (Altschul et al., 1990), which confirmed the uniqueness of the sequence identity of the Opt and SubOpt APC sequences.
PCR using Opt APC (144 repetitions) was compared with that done using infected plant tissue reference positive controls (116 repetitions) by Fisher's exact test (Table 3 ). Nine false negatives occurred among the reference plant positive controls (92% success rate), while the Opt APC was 100% successful (Table 3).
Table 3.
Fisher's exact test (5% significance) comparison of successful PCR amplifications from optimal APC and a reference plant positive control.
| Target | n | Fisher's exact (P-value) |
|
% Opt APC, % reference success rates | |
|---|---|---|---|---|---|
| Opt APC | Referencea | ||||
| BYDV | 190 | 0.0225⁎ | 100/100 | 85/90 | 100%, 94% |
| SBWMV | 16 | 0.2500ns | 12/12 | 3/4 | 100%, 75% |
| TriMV | 9 | 0.2222ns | 7/7 | 1/2 | 100%, 50% |
| WSMV | 45 | 0.1919ns | 25/25 | 18/20 | 100%, 90% |
| Total | 260 | 0.0006⁎⁎⁎ | 144/144 | 107/116 | |
| Total success rate (%) | 100 | 92 | 100%, 92% | ||
ns = no significance.
PCR tests from reference positive controls were from cDNA. All reference positive control tested positive during preliminary assays of this study.
Significant at 0.05.
Significant at 0.001.
3.3. Success of APC with the use of a paper based DNA sampling matrix
Amplification of the BYDV target in Opt APC was successful whether it was blotted onto an EICD matrix (50 mm, 76 mm, 83 mm, or 173 mm) or not. Amplicons obtained from 1.2-mm disks of different soluble paper matrix thickness were of similar band intensity (Supplementary Fig. 2). Also, Fig. 2 shows that the APC control band and the real band were both visible, showing that the intensity of the reaction with APC is comparatively visible with respect to the real band.
Supplementary Fig. 2.
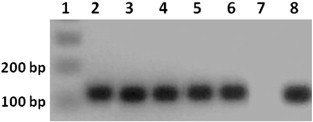
PCR amplification of APC POS-2_7 BYDV positive control embedded in paper based technology prototypes (EICD) of different thickness, a 1.2-mm disk of soluble EICD central membrane was used for PCR. Lane 1 is a 1-Kb plus DNA ladder (Invitrogen, Carlsbad, CA); lane 2 is 83 mm; lane 3 is 76 mm; lane 4 and 5 are 50 mm; lane 6 is 173; lane 7 is an EICD with no plasmid; lane 8 is the optimal APC plasmid only.
4. Discussion
The suitability for PCR of this new concept of customized synthetic DNA positive controls (APCs) consisting of linear arrays of primer sequences designed from diverse genomes or targets or from previously designed primers of value was demonstrated. The performance in PCR of two APCs Opt (POS-2_7) and SubOpt (POS-2_1) designed and constructed after circularization with pUC57 was compared with that of infected plant tissues used as reference positive controls and shown to be more reliable. In fact, APCs with vastly different thermodynamics based on in silico determined ΔG, free energy of major secondary structure and number of predicted secondary structures (Supplementary Table 1) produced visually indistinguishable PCR product yield (Fig. 2), indicating a larger range of thermodynamic difference between these two APCs will be required to influence PCR amplification or product yield. The amount of energy available for a reaction to occur, ∆G, changes as a system moves from its initial state to equilibrium at constant temperature or pressure, therefore, when ∆G is equal to 0, the DNA in the reaction is in equilibrium (Nelson and Cox, 2008). With this assumption, ∆G for unfolding was determined for all 24 APC inserts and the broadest possible ∆G range was selected empirically. An Mfold unfolding plot ∆G of 0.2 kcal mol− 1 and free energy of the major secondary structure − 0.6 kcal mol− 1 were considered to reflect weak interactions among nucleotide bases allowing a more relaxed secondary structure close to the optimal ∆G of 0 kcal mol− 1, while a plot ∆G of 1.0 kcal mol− 1 and free energy of the major secondary structure 1 kcal mol− 1 were considered to reflect less relaxed and stronger nucleotide sub-optimal interactions (Supplementary Table 1) (Jacobson and Zuker, 1993). In this experiment, PCR using Opt APC will be favored since it has a ∆G closer to the equilibrium than SubOpt APC does (Nelson and Cox, 2008). Our results are favorable considering that most APCs will be composed of previously validated primers, which are expected to have a balanced GC content (close to 50%), optimal ∆G (close to zero) and minimal secondary structure. These characteristics should reduce the risk of secondary structure formation over the APC insert. Secondary structure is to be considered during the APC design because it can decrease PCR sensitivity by limiting strand annealing (Jensen et al., 2010). Also, secondary structure in the form of stem-loop structures can lead to polymerase jumping and mispriming during PCR (Jensen et al., 2010, Weiss et al., 1994). The presence of secondary structures within the structure of APC inserts can be easily overlooked while designing by calculating the ∆G of the APC insert in silico, and the number of secondary structure and related free energy before the custom DNA synthesis. In general, the in silico assessment is precautionary because problems associated with folding among primers are not easily resolved by modifying annealing temperature, pH, and salt concentrations at the time of the reaction (Jacobson and Zuker, 1993).
The selected range of plasmid concentration assures the presence of sufficient template for consistent amplification and while plasmid dilutions under 0.1 ng μL− 1 may lead to variable PCR yields. The concentration of a microbial target in a sample is usually unknown, but in an artificial positive control this concentration is known, and has to be sufficient to allow monitoring the performance of the APC during the reaction.
PCR products from APCs may differ in size from those generated by traditionally-used positive controls (Fig. 2) due to the discrepancy in the lengths between the annealing sites of the target sequences in vivo and the arrangement of primer sequences in the APC template (Table 1). Such discrepancy should not be a problem. Certainly the operator knows what APC:sample size product to look for. The generation of products of different sizes by the infected positive control and the APC is appealing to some operators who like the ability to discern the two types of controls as well as to be able to recognize false positives resulting from cross-contamination of APC batches (Charrel et al., 2004, Tao and Zhang, 1998). However, the option for reducing or eliminating the size discrepancy is available. The APC can be designed to have product sizes identical or similar to those expected with reference positive controls (Fig. 2 A) by reducing or adding non complementary nucleotides between primer sets until the desired product size is obtained. Confirmation and quality control can be verified by direct sequencing because each product corresponds to a specific APC sequence identity. Also, the restriction sites flanking the APC could be used to confirm that the correct primer pairs were used in a determination. Alternatively, further monitoring can be performed by high resolution melting (HRM) analysis which allows characterizing each DNA product or APC according to its dissociation behavior (Charrel et al., 2004).
The APC approach described here allows mimicking any kind of target from diverse groups of organisms, including viruses, as well as the inclusion of a large number of targets in one APC or in groups or tandems of APCs (Fig. 1-B). The design in groups or tandems of APCs also brings flexibility to maintain and update a particular group of primers after certain period of time. For example, we can speculate upon an 200 bp long APC insert that harbors one linear array of five different targets with an average of 20 nt per primer (ten primers) (see Fig. 1), from which 120 bp PCR products are amplified (see Table 1). Because the APC sequence is expected to be designed with multiple primer inserts it is unlikely to expect no specific amplifications. Assuming that DNA segments up to 10 kb can be synthesized and cloned into pUC57 using sub-cloning and short DNA oligos as building blocks (Genescript, 2012), an APC insert of 10 kb would carry 50 different sub-APCs in tandem containing 5 primer sets each, which is equivalent to 250 pre-determined microbe targets. Such construct would allow a broad and diverse positive control to include a large number of pre-determined targets for numerous pathogens or genes, eliminating biosafety risks associated to the handling of high consequence pathogens.
When the use of APCs and infected plant tissue was compared in PCR, the former yielded maximum accuracy (100%), while the latter provided only 92% accuracy (Table 3) over all targets (p = 0.0006). For just the BYDV target, the comparison of APCs vs. PCR yielded a difference of 100% to 94% (p = 0.0225). This difference is difficult to attribute to a single factor, and is primarily due to small sample sizes associated with the SBWMV, TriMV and WSMV targets. Actually, the biological significance for SBWMV, TriMV and WSMV is greater than that of BYDV (as evidenced by the greater differences seen in the percentages). This is why the significance for the total is much greater than that of just BYDV (p = 0.0006 vs. p = 0.0225). Loss of accuracy with traditional positive controls is normally associated with RNA or cDNA degradation, human errors occurring during the multi-step extraction, or during reverse transcription, PCR or one step RT-PCR (Chomczynski and Sacchi, 1987, Garber and Yoder, 1983, Park, 2007). Furthermore, in the case of plants, plant-associated PCR inhibitors (polyphenolic compounds, tannins and polysaccharides) can cause anomalous association among PCR components during storage (Li et al., 2008, Murray and Thompson, 1980) and interfere with subsequent assays. This kind of interference was not observed within the APCs PCR amplification because APCs were tested on two-step RT-PCR, which freed APC reactions from host inhibitors making the reaction highly accurate.
The use of plasmid-based PCR controls to mimic pathogens has been reported, but their development is laborious and their impact has been limited to a small number of targets (Caruthers, 1985, Caruthers, 1991, Charrel et al., 2004, Chomič et al., 2010, Inoue et al., 2004, Siegling et al., 1994, Stähler et al., 2006). Advances in their development have included the linear arrangement of several genes linked through restriction and hybridization sites, and the use of a complete gene cloned into a plasmid vector (Charrel et al., 2004, Inoue et al., 2004, Siegling et al., 1994, Smith et al., 2006).
This study described the concept, development and functional validation of fully synthetic APCs, designed and constructed using a novel design involving cloning of custom and de novo synthetic DNA inserts constructed of either primer sequences from multiple genomic targets or existing priming sequences of value for detection and diagnostics. This approach offers the possibility of engineering single controls for a large number of predetermined targets. If adopted, this concept will find applications in many areas of microbiology. Moreover, APC constructs would be used in conjunction with EICD or alternative dry or paper based storage technology to facilitate preservation, shipping and commercialization. The implementation of APCs can eliminate biosafety risks associated with in vivo positive controls, can streamline and improve reliability of PCR detection and diagnostics, and offer new opportunities for development of synthetic positive control combinations to fulfill the demand of different health, forensics or agricultural markets.
The following are the supplementary data related to this article.
ΔG, free energy of major secondary structure and number of predicted secondary structures of 24 APC inserts. Calculations were made by mFOLD at 60˚C, 10 mM Na+ and 1.5 mM Mg++.
Accession numbers incorporated into consensus sequences for primer design.
Funding
This study was supported by Oklahoma State University, Oklahoma Agricultural Experiment Station Project Number OKL02773. The mention of trade names or commercial products in this publication does not imply recommendation or endorsement by Oklahoma State University.
Conflict of interest statement
None declared.
Acknowledgments
The authors acknowledge Drs. Jacqueline Fletcher, Ulrich Melcher and Hassan Melouk, committee members of the doctoral research program of Dr. Donna Caasi at Oklahoma State University, for their mentoring, review and supervision, Jennifer Olson and Dr. Robert Hunger for providing reference plant positive controls, and to Drs. Jacqueline Fletcher and Ulrich Melcher for reviewing this manuscript. The authors also extend their acknowledgment to Oklahoma State University, Oklahoma Agricultural Experiment Station for funding and support.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez A.M. Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu. Rev. Phytopathol. 2004;42:339–366. doi: 10.1146/annurev.phyto.42.040803.140329. [DOI] [PubMed] [Google Scholar]
- Arif M., Ochoa-Corona F.M. Comparative assessment of 5′ A/T-rich overhang sequences with optimal and suboptimal primers to increase PCR yields and sensitivity. Mol. Biotechnol. 2012 doi: 10.1007/s12033-012-9617-5. (First published online on 02 November, 2012.) [DOI] [PubMed] [Google Scholar]
- Caruthers M.H. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- Caruthers M.H. Chemical synthesis of DNA and DNA analogs. Acc. Chem. Res. 1991;24:278–284. [Google Scholar]
- Charrel R., La Scola B., Raoult D. Multi-pathogens sequence containing plasmids as positive controls for universal detection of potential agents of bioterrorism. BMC Microbiol. 2004;4:21. doi: 10.1186/1471-2180-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Chomič A., Pearson M.N., Clover G.R.G., Farreyrol K., Saul D., Hampton J.G., Armstrong K.F. A generic RT-PCR assay for the detection of Luteoviridae. Plant Pathol. 2010;59:429–442. [Google Scholar]
- Fredman D., Jobs M., Strömqvist L., Brookes A.J. DFold: PCR design that minimizes secondary structure and optimizes downstream genotyping applications. Hum. Mutat. 2004;24:1–8. doi: 10.1002/humu.20066. [DOI] [PubMed] [Google Scholar]
- Garber R.C., Yoder O.C. Isolation of DNA from filamentous fungi and separation into nuclear, mitochondrial, ribosomal, and plasmid components. Anal. Biochem. 1983;135:416–422. doi: 10.1016/0003-2697(83)90704-2. [DOI] [PubMed] [Google Scholar]
- Genescript Gene synthesis. 2012. http://www.genscript.com/index.html
- Henson J.M., French R. The polymerase chain reaction and plant disease diagnosis. Annu. Rev. Phytopathol. 1993;31:81–109. doi: 10.1146/annurev.py.31.090193.000501. [DOI] [PubMed] [Google Scholar]
- Inoue S., Noguchi A., Tanabayashi K., Yamada A. Preparation of a positive control DNA for molecular diagnosis of Bacillus anthracis. Jpn. J. Infect. Dis. 2004;57:29–32. [PubMed] [Google Scholar]
- Jacobson A.B., Zuker M. Structural analysis by energy dot plot of a large mRNA. J. Mol. Biol. 1993;233:261–269. doi: 10.1006/jmbi.1993.1504. [DOI] [PubMed] [Google Scholar]
- Jensen M.A., Fukushima M., Davis R.W. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PLoS One. 2010;5:e11024. doi: 10.1371/journal.pone.0011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josue-Caasi D.R. Oklahoma State University; 2012. Assessment of Soluble Biomaterials and Innovation of an Elution-independent Collection Device for Rapid Collection, Detection, And storage of Plant Pathogens; p. 3524481. (Doctoral disertation) [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li R., Mock R., Huang Q., Abad J., Hartung J., Kinard G. A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods. 2008;154:48–55. doi: 10.1016/j.jviromet.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Louw T.M., Whitney S.E., TerMaat J.R., Pienaar E., Viljoen H.J. Oligonucleotide optimization for DNA synthesis. AICHE J. 2011;57:1912–1918. [Google Scholar]
- Menzel W., Jelkmann W., Maiss E. Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. J. Virol. Methods. 2002;99:81–92. doi: 10.1016/s0166-0934(01)00381-0. [DOI] [PubMed] [Google Scholar]
- Miller S.A., Beed F.D., Harmon C.L. Plant disease diagnostic capabilities and networks. Annu. Rev. Phytopathol. 2009;47:15–38. doi: 10.1146/annurev-phyto-080508-081743. [DOI] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechvatal J.M., Ram J.L., Basson M.D., Namprachan P., Niec S.R., Badsha K.Z., Matherly L.H., Majumdar A.P.N., Kato I. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J. Microbiol. Methods. 2008;72:124–132. doi: 10.1016/j.mimet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Nelson P.E., Cox M.M. W.H. Freeman and Company; New York, NY: 2008. Lehninger Principles of Biochemistry. [Google Scholar]
- Ochoa-Corona, F.M., 2012. United States Patent Application US2012/0202211 A1.
- Park D. Genomic DNA isolation from different biological materials protocols for nucleic acid analysis by nonradioactive probes. In: Hilario E., Mackay J., editors. Protocols for Nucleic Acid Analysis by Nonradioactive Probes. Second edition. Vol. 353. Humana Press; New York, NY: 2007. pp. 3–13. (Methods in Molecular Biology). [Google Scholar]
- Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schmith M. Diffusion of synthetic biology: a challenge to biosafety. Syst. Synth. Biol. 2008;2:1–6. doi: 10.1007/s11693-008-9018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegling A., Lehmann M., Platzer C., Emmrich F., Volk H.-D. A novel multispecific competitor fragment for quantitative PCR analysis of cytokine gene expression in rats. J. Immunol. Methods. 1994;177:23–28. doi: 10.1016/0022-1759(94)90139-2. [DOI] [PubMed] [Google Scholar]
- Smith G., Smith I., Harrower B., Warrilow D., Bletchly C. A simple method for preparing synthetic controls for conventional and real-time PCR for the identification of endemic and exotic disease agents. J. Virol. Methods. 2006;135:229–234. doi: 10.1016/j.jviromet.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stähler P., Beier M., Gao X., Hoheisel J.D. Another side of genomics: synthetic biology as a means for the exploitation of whole-genome sequence information. J. Biotechnol. 2006;124:206–212. doi: 10.1016/j.jbiotec.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Tao Q., Zhang H.-B. Cloning and stable maintenance of DNA fragments over 300 kb in Escherichia coli with conventional plasmid-based vectors. Nucleic Acids Res. 1998;26:4901–4909. doi: 10.1093/nar/26.21.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J., Zucht H.D., Forssmann W.G. Amplification of gene fragments with very high G/C content: c7dGTP and the problem of visualizing the amplification products. Genome Res. 1994;4:124–125. doi: 10.1101/gr.4.2.124. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ΔG, free energy of major secondary structure and number of predicted secondary structures of 24 APC inserts. Calculations were made by mFOLD at 60˚C, 10 mM Na+ and 1.5 mM Mg++.
Accession numbers incorporated into consensus sequences for primer design.


