Fig. 2.
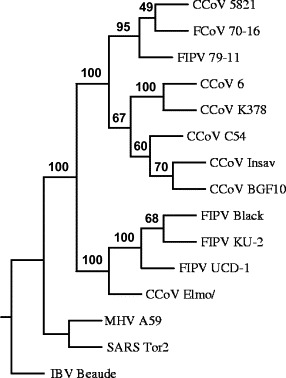
Phylogenetic tree of spike protein of canine and feline coronaviruses. Phylogenetic tree was constructed by the protein sequence parsimony method (protpars) using Phylip 3.6 through the Phylogeny Interface Environment (PIE) at the Rosalind Franklin Centre for Genomics Research. Bootstrap values were calculated on 1000 replicates randomising the input order 41 times. The tree was rooted with MHV A59, SARS Tor2 and IBV Beaudette. FIPV, feline infectious peritonitis virus; FCoV, feline coronavirus; CCoV, canine coronavirus; MHV, mouse hepatitis virus; SARS, severe acute respiratory syndrome; IBV, infectious bronchitis virus.
