Fig. 1.
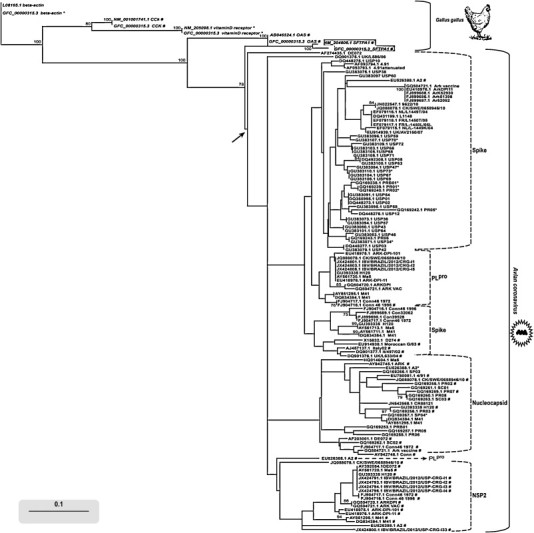
Neighbor-joining distance tree for the relative synonymous codon usage (RSCU) for the Avian coronavirus spike (S), nucleocapsid (N), non-structural protein 2 (NSP2) and papain-like protease (PLpro) genes and the Gallus gallus beta-actin, lung surfactant protein A (SFTPA1, gray background), intestinal cholecystokinin (CCK), oviduct ovomucin alpha subunit (OSA) and kidney vitamin D receptor genes. The tree was based on binary data using the value 1 for RSCUs > 1 (codon is preferred) or 0 for RSCUs ≤ 1 when the codon is not preferred (RSCU < 1) or is neutral (RSCU = 1). ENC (effective number of codons) values <40 and >45 are marked with an asterisk and a hash, respectively; sequences with ENC values between 40 and 45 have no marks. The arrow indicates the separation between G. gallus and Avian coronavirus clusters. Numbers at each node are bootstrap values (1000 replicates, only values >50 are shown). The bar represents the codon usage preferences distance.
