Fig. 5.
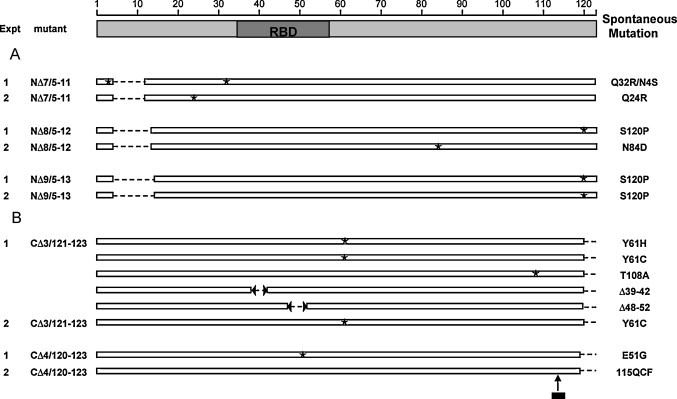
Spontaneous mutations in the N protein of recovered P5 N- and C-terminal mutant viruses. Schematic representation of N protein amino acid sequence is shown at the top; RBD indicates the predicted RNA binding domain, while NLS-1 and NLS-2 represent the regions of two nuclear localization signals. The experiment (Expt) 1 and 2 indicates the independent transfection assays. The corresponding nucleotide changes are described in the text. (A) Spontaneous mutations in the N-terminal deletion mutant viruses. The positions of the engineered deletions (dashed lines) and the spontaneous substitutions (asterisks) are described relative to the schematic representation. (B) Spontaneous mutations in the C-terminal deletion mutant viruses. Dashed lines indicate the engineered deletions, whereas the asterisks represent substitutions, the broken arrows indicate sequence deletions, and the short bold line indicates the 3-aa insertion. 115QCFRVTA in experiment 2 of mutant CΔ4/120–123 indicates the fully changed final seven residues, which resulted from the 9-nt insertion between 15341T and 15342T.
