Fig. 2.
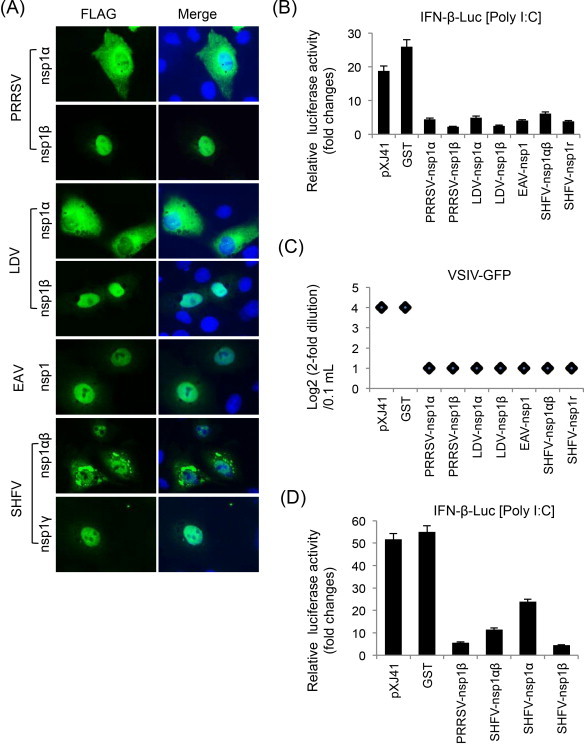
Suppression of IFN-β production by individual subunits of nsp1 of arteriviruses. (A) Subcellular localization of individual nsp1 subunits of arteriviruses in MARC-145 cells. Cells were grown to 40% confluency and transfected with indicated genes for 24 h. Cells were stained with anti-FLAG Ab followed by staining with Alexa 488-labeled anti-mouse Ab and DAPI. Cellular localization of individual subunits was examined by fluorescent microscopy. (B) HeLa cells were seeded in 12-well plates and co-transfected with pIFN-β-Luc along with individual genes and pTK-RL as an internal control at a ratio of 1:1:0.1. At 24 h post-transfection, cells were stimulated with 1 μg/ml of poly(I:C) for 12 h, followed by lysis and determination for reporter activity using the dual luciferase assay system (Promega). Relative luciferase activities were calculated by normalizing the firefly luciferase to renilla luciferase according to the manufacturer's protocol. The data represent the means of three independent experiments, each experiment in triplicate. (C) IFN bioassays using VSIV-GFP (vesicular stomatitis Indiana virus expressing GFP). HeLa cells in 6-well plates were transfected with individual genes for 24 h, and stimulated with poly(I:C) for 12 h. Cell culture supernatants were collected and diluted serially by twofolds. MARC-145 cells were grown in 96-well plates and incubated with each dilution of supernatants for 24 h, and then infected with VSIV-GFP at an MOI of 0.1 for 16 h. VSIV replication was measured by monitoring the fluorescence by GFP expression. (D) Identification of the functional region for IFN-β suppression in SHFV-nsp1αβ. SHFV-nsp1α and SHFV-nsp1β were separated according to Vatter et al. (2014) to make SHFV-nsp1α as 164 aa in length and SHFV-nsp1β as 186 aa.
