Fig. 6.
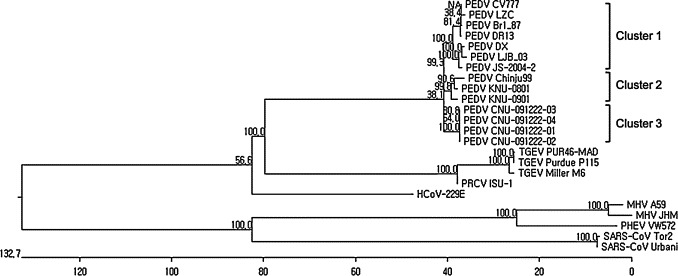
Phylogenetic analysis based on the amino acid sequences. The phylogenetic tree was generated using the Clustal W method following multiple alignments of amino acid sequences. Putatively similar regions of spike proteins of other distantly related coronaviruses were also included in this tree. The numbers represent bootstrap values greater than 50% of 1000 replicates. New PEDV isolates were in different cluster from those published.
