Fig. 2.
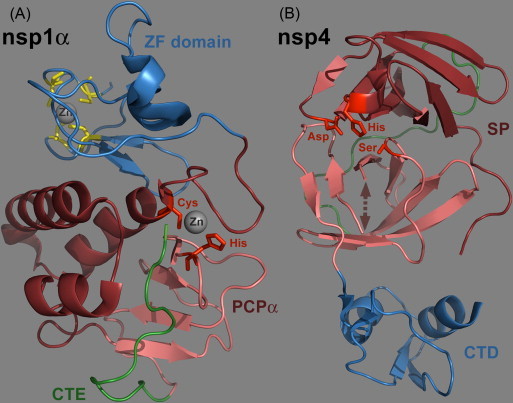
Ribbon diagrams of the PRRSV nsp1α and nsp4 crystal structures. Representations were made using the Pymol molecular graphics system (DeLano, 2002) and PDB entries 3IFU (Sun et al., 2009a) and 3FAN (Tian et al., 2009), respectively. (A). Crystal structure of an nsp1α monomer consisting of five β strains and five α helices (Sun et al., 2009a). The N-terminal ZF domain (Met1 to Glu65; Zn-binding residues highlighted in yellow) and C-terminal PCPα domain (Pro66 to Gln166) are colored as blue and dark red ribbons, respectively. The ‘C-terminal extension’ (CTE: Arg167 to Met 180) that resides inside the PCPα substrate-binding pocket is featured in green. (B). Crystal structure of PRRSV nsp4 (Tian et al., 2009). The protein consists of three domains of which the first two (in dark red and pink) form the typical chymotrypsin-like two-β-barrel fold. A long loop connecting domains I and II is highlighted in green, whereas the arrow represents a loop consisting of residues 136–140, which was devoid of visible electron density. The residues of the catalytic triad are highlighted in red. The C-terminal domain was shown to be dispensible for proteolytic activity in EAV nsp4 and is depicted in blue (Barrette-Ng et al., 2002, van Aken et al., 2006b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
