Fig. 1.
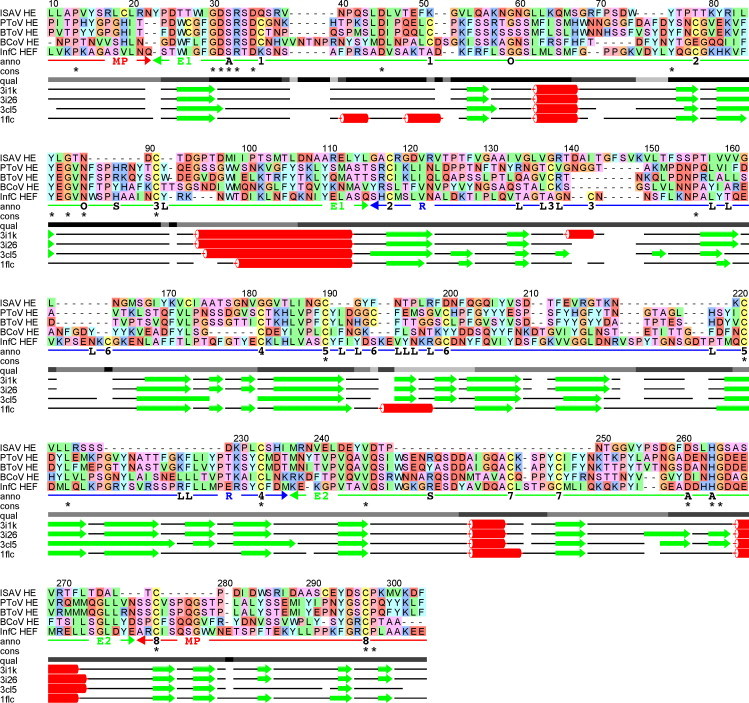
Multiple alignment of hemagglutinin sequences, showing the FFAS (Rychlewski et al., 2000) alignment of ISAV HE (Q911Y4) against the structure-based alignment of porcine torovirus HE (PToV HE; PDB id code 3I1K), bovine torovirus HE (BToV HE; 3I26), bovine coronavirus HE (BCoV HE; 3CL5) and influenza C virus HEF (InfC HEF; 1FLC). The upper part shows the sequence alignment, the lower part shows the corresponding alignment of secondary structure elements determined from the experimental protein structures (β-strands: green arrows, α-helices: red coils). The qual line indicates alignment quality based on the degree of consensus between FFAS alignments against each of the structural templates, ranging from full consensus in black to no consensus in light grey. The cons line shows completely conserved positions in the alignment. The anno line shows relevant annotation of the alignment, including residues involved in active site (A), oxyanion hole (O), ligand binding (L), substrate coordination (S) and Cys–Cys pairing (1–8). The anno line also shows domain organisation (MP: membrane-proximal (red); E1, E2: esterase (green); R: receptor (blue)) taken from Langereis et al. (2009). The receptor domain is frequently annotated as a lectin domain in other publications, e.g. in Langereis et al. (2009). The numbering of the alignment is according to the full-length ISAV HE sequence. The alignment includes part of the signal peptide for ISAV HE (LLAPVYS from position 10 to 16), but no part of the signal peptide for any of the other sequences. This shows a clear sequence similarity between part of the ISAV HE signal peptide and the structural templates.
