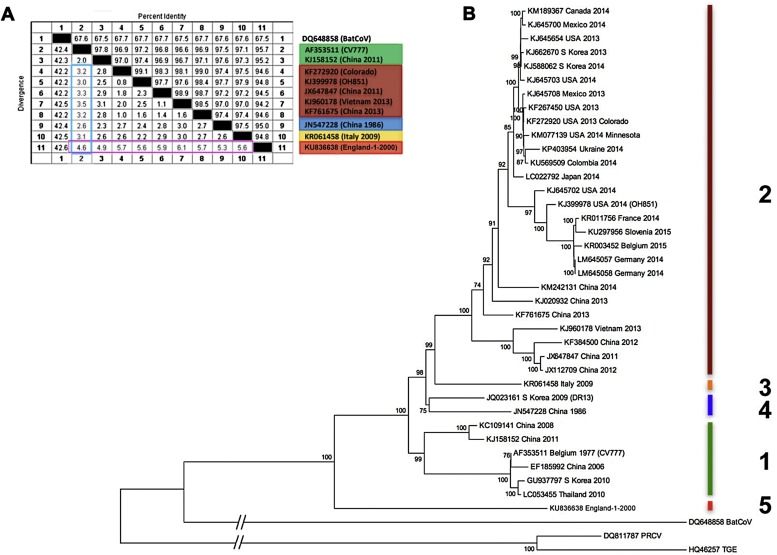
Fig. 1.
Phylogenetic analysis of 40 representative PEDV full genome RNA sequences. The evolutionary history was inferred by using the Neighbor-Joining method, as described by Huang et al., 2013. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). Accession numbers, country and year of isolation are indicated, as provided by GenBank and the references therein. The resulting genotypes are numbered and colour-coded. A) Percentage identity/differences across the different genotypes. The genetic differences between genotypes are at least 2.5% as highlighted in the light blue box using the prototype PEDV strain CV777 as reference. This table also demonstrates that PEDV England-1-2000 (Genotype 5) is more diverse to any other genotype than these are to each other (>4.5%, pink box). B) The tree is drawn to scale, except for the out-groups bat coronavirus (BatCoV), porcine respiratory coronavirus (PRCV) and transmissible gastroenteritis virus (TGEV), which are shown for reference. Branch lengths other than the out-groups reflect the number of substitutions per site. The percentage of trees in which the associated taxa clustered together is shown next to the branches for values greater than 70%. Genotypes 1 and 2 are as inferred by Huang et al. (2013); Genotype 3 is represented by the recently sequenced Italian strain from 2009, while Genotype 4 is represented by two strains derived from recombination thus previously termed R (Huang et al., 2013). Genotype 5 is represented by the strain England-1-2000, the most distinct PEDV strain discovered so far.