Fig. 3.
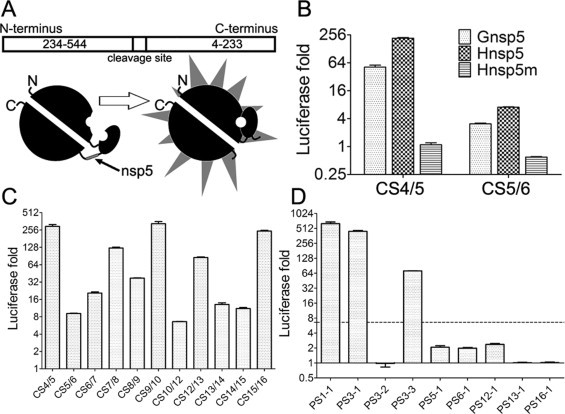
Identification of the cleavability of predicted cleavage sites in recombinant luciferase cleavage assays. (A) Schematic diagram of the recombinant luciferase. (B) Verification of the recombinant luciferase assays. Inactive luciferase was synthesized in the cell-free translation system and the reaction mixture incubated at 25 °C for 2 h. After that, the protein mixture was divided into four parts and incubated with 1.63 μM Gnsp5, Hnsp5, Hnsp5m or H2O, respectively. After incubation for 1 h at 30 °C, the reaction product was diluted 20 times and mixed with equal amount of luciferase substrate. After incubation at room temperature for 5 min, the luciferase luminescence was measured. Luciferase activation fold was calculated through dividing the signal value of the reaction system treated with active Hnsp5 by the one treated with the inactive nsp5 mutant Hnsp5m. (C) The luciferase cleavage assay of predicted 11 canonical cleavage sites and (D) 9 putative cleavage sites. The luciferase expression vector inserted with cleavage sites were added to the wheat germ protein translation mix and incubated at 25 °C for 2 h, and the reaction mixture was divided and treated with Hnsp5 and Hnsp5m, respectively. The dashed line indicates the lowest fold increase of luciferase signal by cleavage of previously confirmed 3CLpro cleavage sites. The data presented here are the mean values ± SD derived from three independent experiments.
