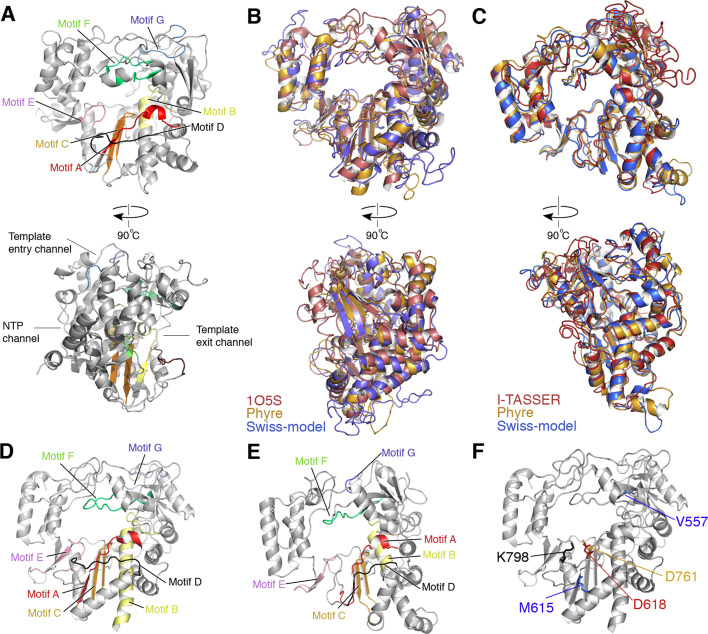
Fig. 3.
Models of the nidovirus RdRps. A) Structure of the FMDV RdRp (pdb 2E9R). In the top panel, conserved polymerase motifs A-F are indicated. In the bottom panel, the template entry, template exit and NTP entry channels are indicated. B) Models of the SARS-CoV nsp12 RdRp generated by Swiss-Model and Phyre2 superposed on the model 1O5S (Xu et al., 2003). All three models are based on the C-terminal polymerase domain and excluded the N-terminal NiRAN domain. All models show an overall similar fold. Differences exist in the fingers subdomain and surface loops of the thumb subdomain. C) Superposed models of the EAV nsp9 RdRp generated by use of I-TASSER, Swiss-Model and Phyre2. Overall, all three models show a very similar fold, with small differences in the fingers subdomain. D) Motifs of the polymerase domain of SARS-CoV nsp12 indicated on the structural model 1O5S (Xu et al., 2003). E) Motifs of the polymerase domain of EAV nsp9 that was generated using Phyre2. F) Key active site and fidelity residues indicated on the model of polymerase domain of SARS-CoV nsp12 1O5S.