Fig. 1.
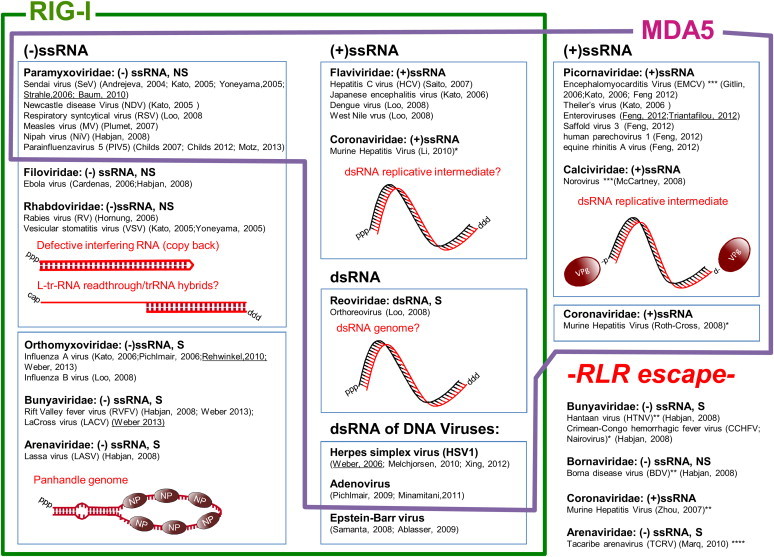
Viruses recognized by RIG-I and MDA5 and evading RLR recognition. S: segmented, NS: non-segmented, ssRNA: single-stranded RNA, dsRNA: double-stranded RNA, (+): positive strand genome, (−): negative strand genome. *Recognition by RIG-I and MDA5 in oligodendrocytes (Li et al. 2010) but no recognition by RIG-I or MDA5 in BM-DC or fibroblasts (Zhou and Perlman 2007), recognition by MDA5, not RIG-I in macrophages and microglia (Roth-Cross et al. 2008). **Evasion of RIG-I recognition by nuclease 5′ end cleavage leaving monophosphate at the 5′end of the viral genome (Garcin et al., 1995, Habjan et al., 2008). ***Evasion of RIG-I recognition via substitution of 5′triphosphate by Vpg protein at the 5′end of the viral genome (Hruby and Roberts, 1978, Lee et al., 1977, Rohayem et al., 2006). ****Evasion of RIG-I recognition by overhang at the 5′end of the viral genome (Marq et al. 2010b). Weber, 2013: papers contributing to the characterization of the real ligand structure in vivo are underlined.
