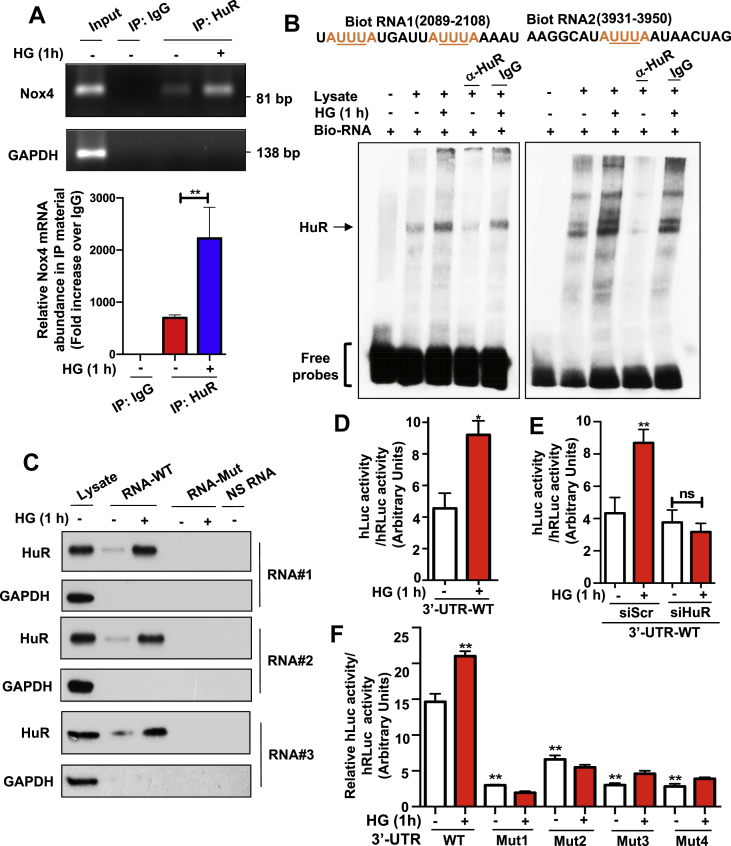
Figure 3.
Transcripts encompassing the Nox4 3′-UTR associate with HuR protein and the abundance of the complexes increased in HG-treated MCs. (A) Nox4 mRNA was presented in the immunoprecipitants with HuR antibody in MCs, and its affinity was increased when cells were exposed to HG. Standard IP protocol was followed. Nox4 mRNA was detected by RT-PCR (upper panel) and quantified against the trace amount presented in total IgG complex (bottom panel). GAPDH served as a control. (B) Nox4 promoter containing AREs motifs were associated with HuR protein. RNA probes were synthesized with biotinylating labeling and incubated with MCs in the presence or absence of the HuR antibody. The RNA EMSA experiments were performed following the LightShift Chemiluminescent RNA EMSA Kit from ThermoSci Inc. (C) HuR protein was presented in RNA pull-down complex withwild type Nox4 promoter RNAs but not with mutated Nox4 promoter RNAs. Mutations were generated on ARE motifs located within Nox4 promoter RNAs. NS-RNA were random, non-specific binding RNAs that were used as negative controls. RNA pull-down complex was probed with the HuR antibody. (D–F) MCs were transiently transfected with a pEZX-MT02-hNox4 3′-UTR construct, a luciferase reporter construct containing human Nox4 3′-UTR region, and luciferase activity was recorded in various conditions. HG increased the luciferase reporter activity in MCs that were transiently transfected with this construct (D). Such effects were reduced by co-transfection with siRNA against HuR in HG-treated MCs (E). Mutation on ARE regions of Nox4 3′-UTR within pEZX-MT02-hNox4 3′-UTR construct disrupted the increased luciferase activity in MCs that were treated with HG (F). All data are presented as the mean ± S.E of three repeats. ∗, p < 0.05; ∗∗, p < 0.01 in comparison with control or as indicated. The Student's t-test was used to compare the two groups; and one-way ANOVA was used for comparison between multiple groups.