Figure 2.
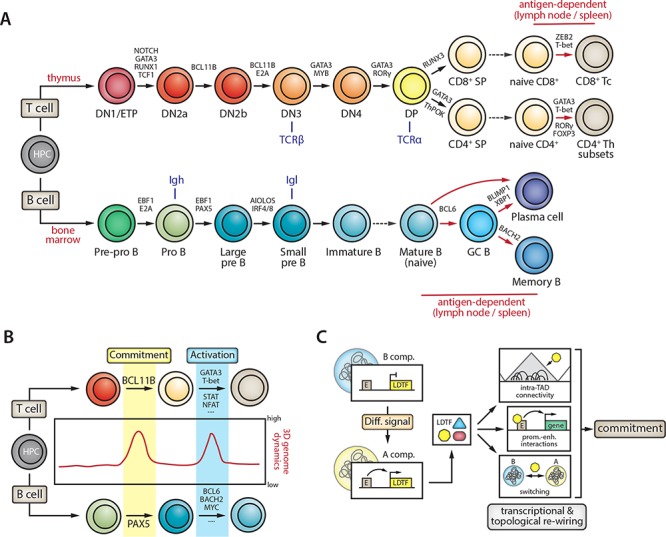
Transcription factors drive 3D genome dynamics during lymphocyte differentiation and activation. (A) Differentiation stages of T (top; for the most common αβ subset) or B (bottom) lymphocytes as they develop from hematopoietic progenitor cells (HPCs) into mature antigen-receptor-expressing cells that migrate to peripheral tissues (i.e. lymph nodes and spleen) for their antigen-dependent activation. Selected key transcription factors (TFs) critical for the various cell state transitions are indicated. Timing of TCR and immunoglobulin (Ig) locus recombination is indicated in blue. Dashed dark gray arrows denote migration of differentiated cells into the periphery; red arrows are used for antigen-dependent differentiation steps. Additional abbreviations: DN, double-negative; DP, double-positive; SP, single-positive; ETP, early thymic precursor; Th, T helper cell; Tc, cytotoxic T cell; GC, germinal center. (B) Analyses of topological genome dynamics during T-cell or B-cell lineage commitment and subsequent activation of mature lymphocytes have shown that the most extensive rewiring of 3D genome organization occurs right after the appearance of the commitment TFs BCL11b (for T cells) or PAX5 (for B cells) and after antigen-dependent activation. Several TFs critical for T-cell or B-cell activation have also been implicated in reorganizing lymphocyte genome conformation (e.g. GATA3, MYC). (C) Schematic representation of the interplay between genome topology, the activation of lineage-determining TFs (LDTF) and the subsequent action of these LDTFs. In the example, the locus encoding an LDTF (e.g. BCL11B in early T-cell development) is switched from the repressive B compartment to the transcription-competent A compartment under the influence of differentiation (Diff.) signals. As the LDTF gene is activated in the A compartment, TF proteins are produced that initiate a transcriptional and topological rewiring of the lymphocyte precursor that will eventually result in stable lineage commitment. LDTFs operate at different levels of 3D genome organization, including modifications to intra-TAD connectivity, promoter–enhancer (prom.-enh.) interactions and A/B compartment switching.
