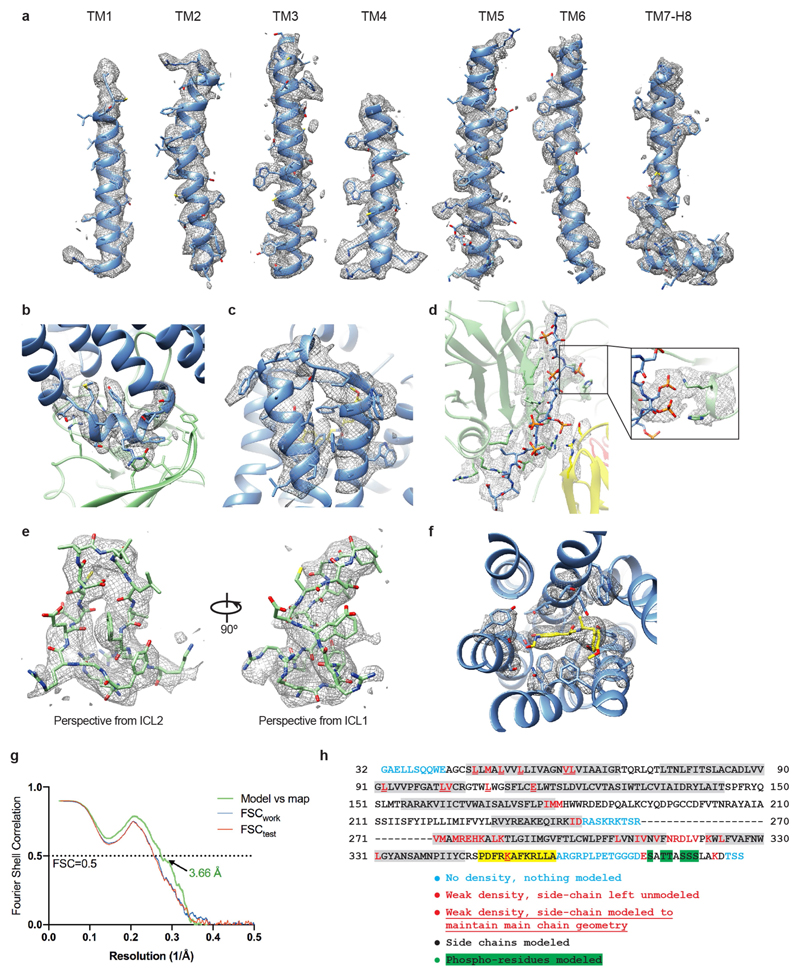
Extended Data Fig. 5. Cryo-EM map quality of the β1AR6P–βarr1–Fab30 complex and model validation.
Unless otherwise stated, density maps were visualized using Chimera38 (contour level 0.017) and encompass a radius of 2 Å around the region of interest. a, Transmembrane helices of β1AR6P with density shown as a mesh. b, ICL2 of β1AR6P. For clarity, the neighbouring βarr1 side chains are depicted without density. c, ECL3 of β1AR6P and the adjacent helical turns of H6 and H7. d, The phosphorylated V2R6P C terminus. Inset, interaction between the V2R6P phospho-threonine dyad and the βarr1 lariat loop. Density in the inset is depicted with contour level 0.01 (carve radius 2 Å). e, The finger loop of βarr1. f, Formoterol and the neighbouring side chains in the orthosteric binding site. g, FSC of the refined model versus the map (green curve), and FSCwork and FSCfree validation curves (blue and red curves, respectively). h, Amino acid sequence of the β1AR6P construct used for the cryo-EM structure determination. The residues are numbered according to the wild-type sequence of β1AR. Residues are coloured according to how they have been modelled (key). Regions highlighted in grey represent the transmembrane α-helices, amphipathic helix 8 is highlighted in yellow, and phosphorylated residues are highlighted in green. The dashes represent amino acid residues that were deleted.