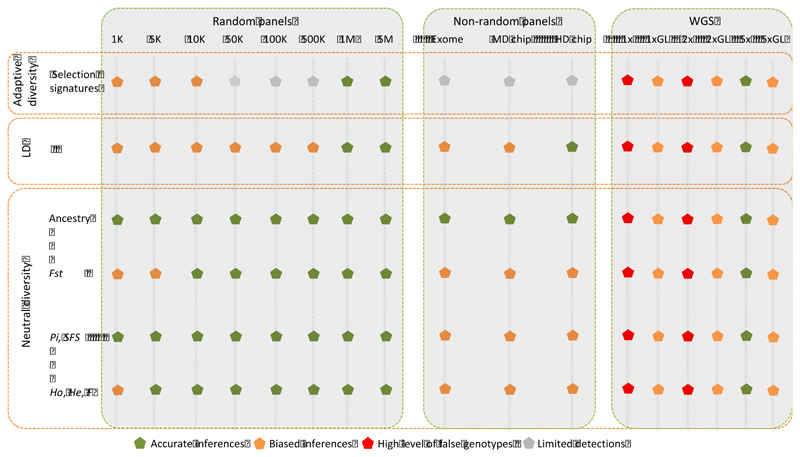
Figure 8. Efficiency and accuracy of different genotyping strategies.
For each purpose, the different strategies are rated according to the accuracy of the estimates taking as a reference the WGS 12x depth inferences. Grey dots indicate that the genotyping approach allow detecting some selection signatures but could miss some further signals detected by high density panels and WGS (12x depth) data. MD chip = 50K SNP BeadChip (caprine and ovine); HD chip = 600K SNP Ovine BeadChip. Low and medium re-sequencing coverages are represented by: (i) classical variant calling and filtering denoted by 1x, 2x and 5x and (ii) variant discovery based on genotype likelihoods denoted by 1xGL, 2xGL and 5xGL.