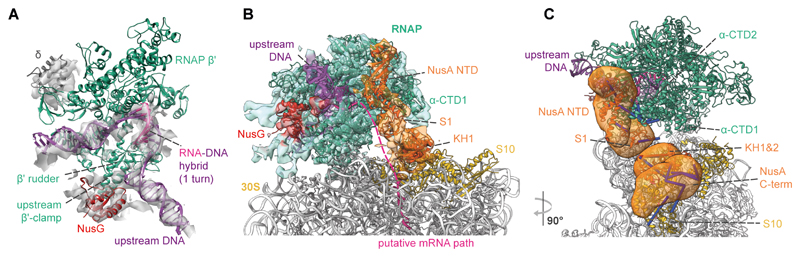
Fig. 3. Integrative model of the M. pneumoniae elongating expressome.
(A) Cryo-EM density of RNAP corresponding to the DNA, RNA-DNA hybrid, upstream β’ clamp, δ subunit and NusG, accommodates one turn of the RNA-DNA hybrid consistent with an elongating RNAP. (B) Integrative model with cryo-EM density of the RNAP-NusG-NusA-ribosome elongating expressome. Structured regions are represented as colored cartoons. The electron density colors represent the subunits occupying the corresponding volumes. Coarse-grained regions are not shown. Schematic of the putative mRNA path refers to the shortest distance between mRNA exit and entry sites. (C) The mRNA exit tunnel face of RNAP is covered by NusA. Localization probability densities for NusA domains are shown in orange. Crosslinks between NusA and other proteins are shown. Satisfied crosslinks (<35 Å) are in blue, overlong crosslinks are in red.