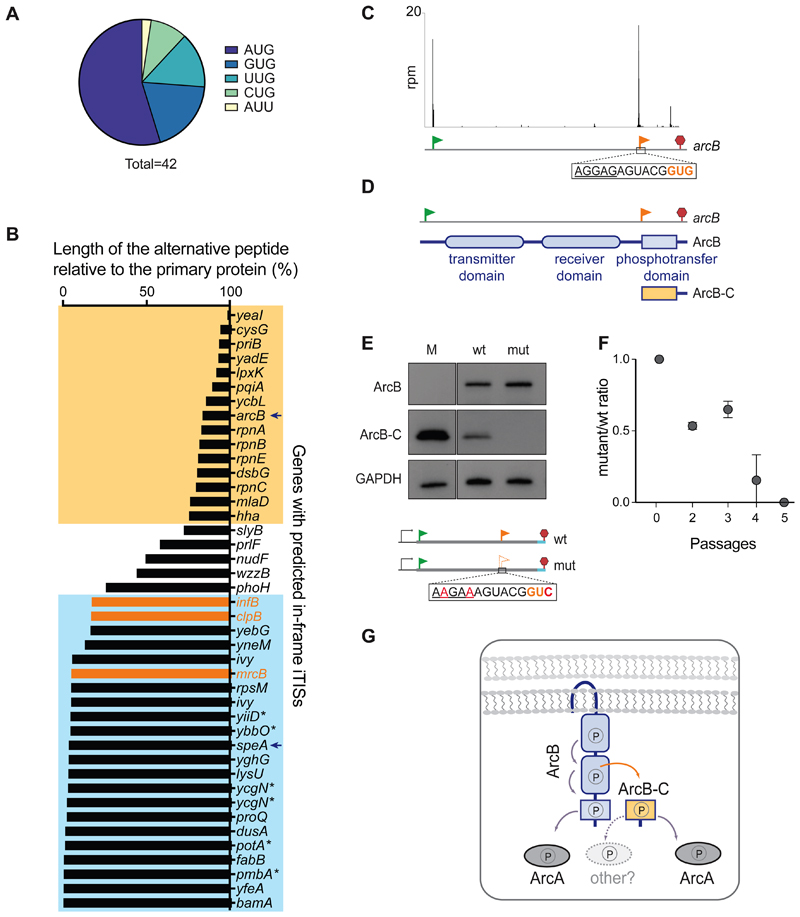
Figure 3. In-frame internal initiation can generate functional N-terminally truncated proteins.
(A) The frequency of various putative start codons at the in-frame iTISs. (B) The relative length of the predicted alternative proteins, products of internal initiation, in comparison with the main protein. The known examples of genes with in-frame iTISs (Figure 2B) are in orange. The genes with the iTISs located within the 3’ or 5’ quartile of the gene length are boxed in yellow or blue, respectively. Asterisks show genes with pTIS re-annotation proposed based on TET-assisted Ribo-seq, (Nakahigashi et al., 2016). Arrows point at arcB and speA genes further analyzed in this work.
(C) Ribo-RET profile of arcB showing ribosomal density peaks at the pTIS (green flag) and the iTIS (orange flag).
(D) Schematics of the functional domains of ArcB. The putative alternative ArcB-C protein would encompass the phosphotransfer domain.
(E) Western blot analysis of the C-terminally 3XFLAG-tagged translation products of the arcB gene expressed from a plasmid in E. coli cells. Inactivation of iTIS by the indicated mutations (mut) abrogates production of ArcB-C. Lane M: individually-expressed marker protein ArcB-C-3XFLAG (see STAR Methods).
(F) Functional iTIS in arcB facilitates growth under low oxygen conditions. BW25113 ΔarcB E. coli cells, expressing from a plasmid either wt arcB or mutant arcB with inactivated iTIS, were co-grown in low oxygen conditions and the ratio of mutant to wt cells was analyzed (see Start Methods for details). Error bars represent deviation from the mean (n=2).
(G) The phosphorelay across the wt ArcB domains results in the activation of the response regulator ArcA (Alvarez et al., 2016). Diffusible ArcB-C could amplify the signal capabilities of the ArcBA system and/or enable cross-talk with other response regulators.
See also Figure S2.