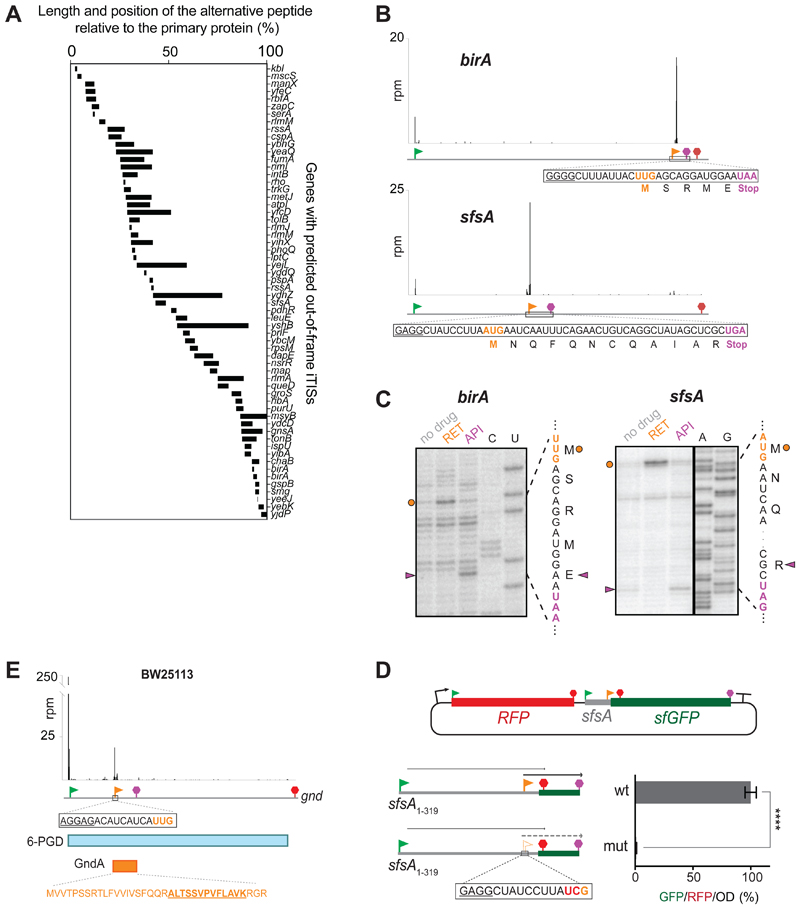
Figure 4. OOF iTISs revealed by Ribo-RET can direct initiation of translation.
(A) The length and location of the alternative OOF protein-coding segments relative to the main ORF.
(B) Ribo-RET profiles of birA and sfsA genes with putative OOF iTISs. The iTIS OOF start codon and the corresponding stop codon are indicated by orange flag and stop sign, respectively. The sequences of the alternative ORFs are shown. (C) Toeprinting analysis reveals that RET arrests ribosomes at the start codons (orange circles) of the alternative ORFs within the birA and sfsA genes; termination inhibitor Api137 arrests translation at the stop codons (purple arrowheads) of those ORFs. The nucleotide and amino acid sequences of the alternative ORFs are shown. Sequencing lanes are indicated.
(D) In the cell, translation is initiated at the sfsA OOF iTIS. Schematics of the RFP/sfGFP reporter plasmid. The rfp and sf-gfp genes are co-transcribed. The sfGFP-coding sequence is expressed from the iTIS (orange flag) and is OOF relative to the sfsA pTIS start codon (green flag). The first stop codon in-frame with the pTIS (red) and the stop codon in-frame with the iTIS (orange) are indicated. The bar graph shows the change in relative green fluorescence in response to the iTIS start codon mutation (mut). The values represent the mean ± standard deviation from technical replicates (n=6). Two-tailed unpaired t-test. (E) Ribo-RET snapshots of the gnd gene revealing the putative location of the start codon of the alternative ORF. The amino acid sequence of the alternative ORF product GndA is shown in orange; the tryptic peptide identified by mass spectrometry (Yuan et al., 2018) is underlined.
See also Figure S5 and Table S1.