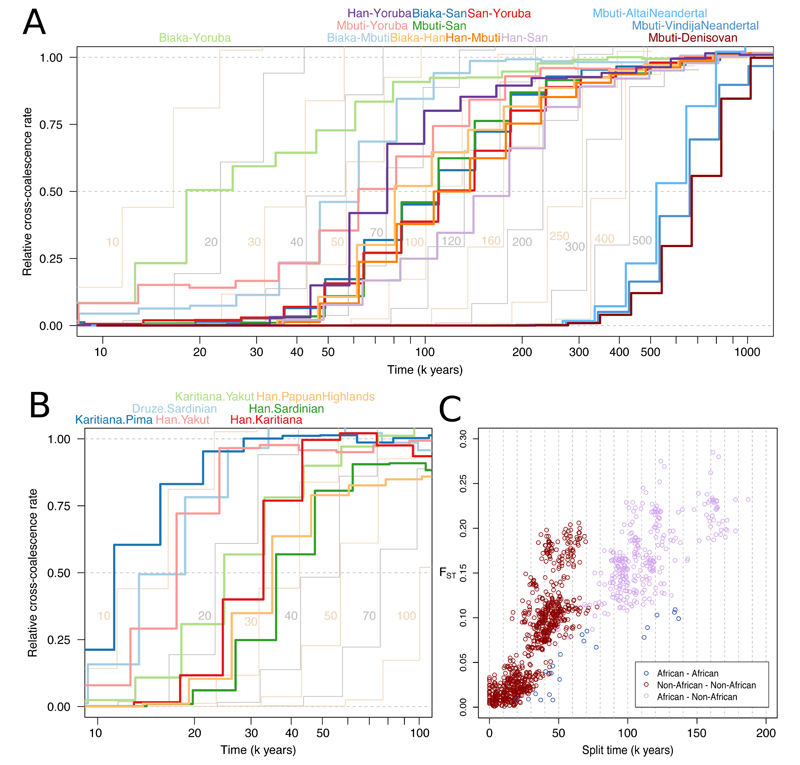
Figure 5. The time depth and mode of population separations.
(A) MSMC2 cross-population results for pairs of African populations, including Han Chinese as a representative of non-Africans, as well as between archaic populations and Mbuti as a representative of modern humans. Curves between modern human groups were computed using four physically phased haplotypes per population, while curves between modern and archaic groups were computed using two haplotypes per population and unphased archaic genomes. The results of simulated histories with instantaneous separations at different time points are displayed in the background in alternating yellow and grey curves. (B) MSMC2 cross-population results, as in A, for pairs of non-African populations. (C) Split times estimated under simple, sudden pairwise split models using momi2 for all possible pairs among the 54 populations against FST, a measure of allele frequency differentiation. The plot does not include Native American populations, as we could not obtain reliable momi2 fits for these.