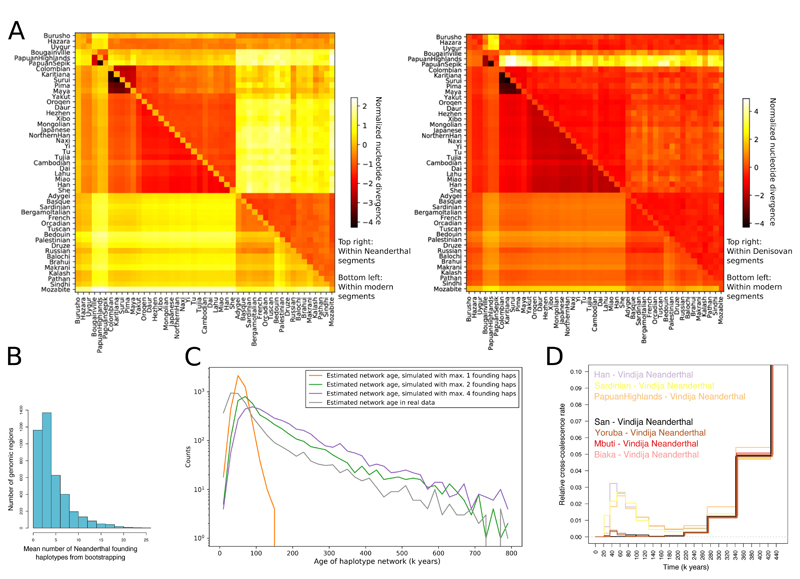
Figure 6. Archaic haplotypes in modern human populations.
(A) Nucleotide divergence DXY within segments deriving from archaic admixture and within other segments in non-African populations. (B) The mean number of archaic founding haplotypes estimated by constructing maximum likelihood trees for each archaic segment identified in present-day non-Africans, and then determining the number of ancestral branches in the tree at the approximate time of admixture (2000 generations ago). (C) The distribution of estimated ages of archaic haplotype networks in the present-day human population. The distribution is compared to results obtained in simulations performed with different numbers of archaic founding haplotypes. (D) MSMC2 cross-population results for African (two individual curves per population) and selected non-African (one individual curve per population) against the Vindija Neanderthal, zooming in on the signal of Neanderthal genome flow in modern human genomes (note the highly reduced range of the vertical axis).