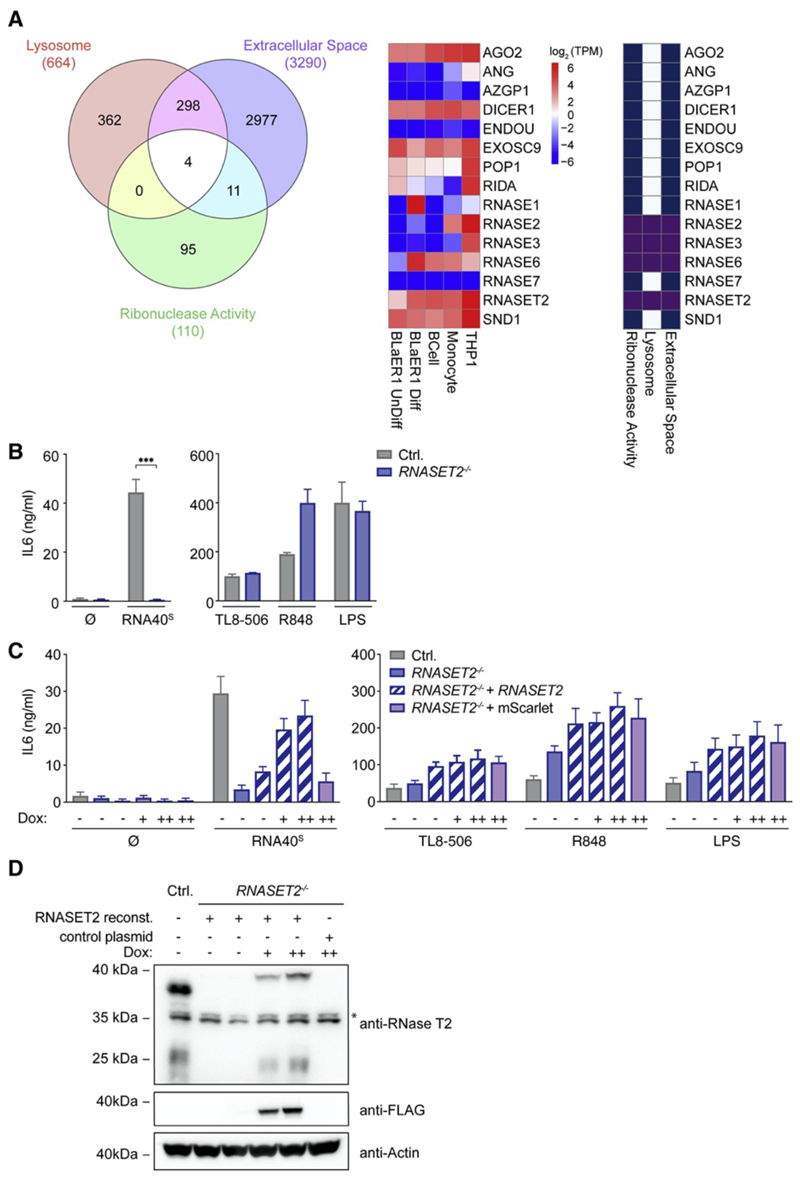
Figure 2. RNase-T2-Deficient Cells Fail to Respond to RNA Oligonucleotides.
(A) Venn diagram of proteins that are, according to their Gene Ontology (GO) terms, annotated as “lysosomal,” “extracellular space,” or “ribonuclease activity” (left). A heatmap shows RNA expression levels of several RNases and their GO-term designation in the indicated cell types (middle and right). (B) IL-6 production in BLaER1 Controls and RNASET2 −/− cells stimulated with RNA40S, TL8-506, R848, and LPS for 14 h. (C) RNASET2 −/− cells were reconstituted with doxycycline-inducible RNase T2 (++, 1 μg/mL; +, 0.5 μg/mL). An inducible mScarlet construct was used as a control. (D) The immunoblot corresponding to the reconstitution experiment is shown. The asterisk indicates unspecific bands. Data are depicted as mean + SEM of three independent experiments or one of three representative blots. Statistics indicate significance by two-way ANOVA: ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05; ns, not significant. See also Figure S2.