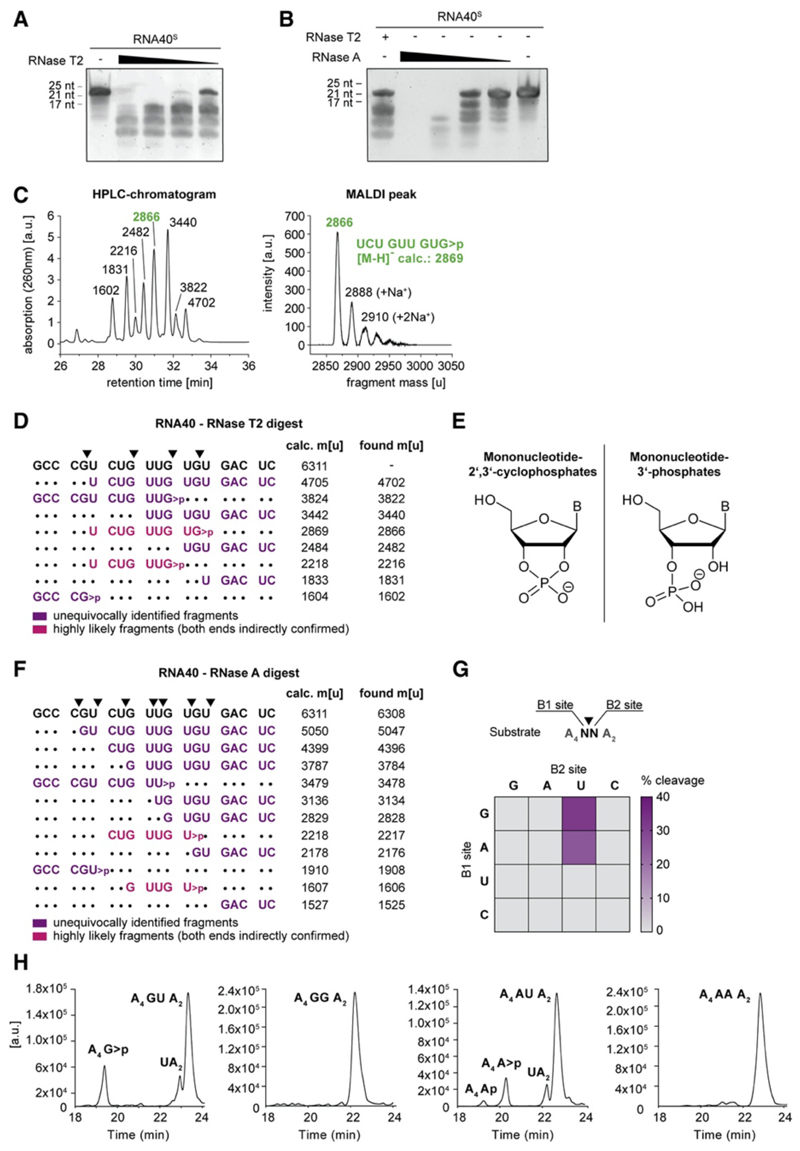
Figure 3. RNase T2 Cleaves RNA between Purine Bases and Uridine.
(A and B) Urea gel of RNA40S digested with decreasing RNase T2 (A) or RNase A (B) concentrations. One representative gel of two independent experiments is shown. (C) HPLC chromatogram of RNA40O-derived ON fragments (left; fragment masses determined by MALDI-TOF) and the corresponding MALDI peak of one representative peak (right) (D) RNA40O was digested with RNase T2 and analyzed by HPLC and MALDI-TOF. The most likely assigned fragments based on MALDI-TOF analysis are depicted next to all calculated and found masses. Calculated masses are shown as [M-H]−. (E) Structure of mononucleotide 2ʹ,3ʹ-cyclophosphates (left) and Mononucleotide 3ʹ-phosphates (right) (F) RNA40O was digested in vitro with RNase A and analyzed as above. Masses are shown as [M-H]−. (G) 16 ONs containing all possible dinucleotide combinations (A4NNA2) were analyzed after RNase T2 in vitro digestion. Of note, all ONs with a U at the B1 site were also cut between A and U, whereas cleavage between U and N was not detected. (H) HPLC chromatograms corresponding to the in vitro digests of GU, GG, AU, and AA (G). See also Figure S3.