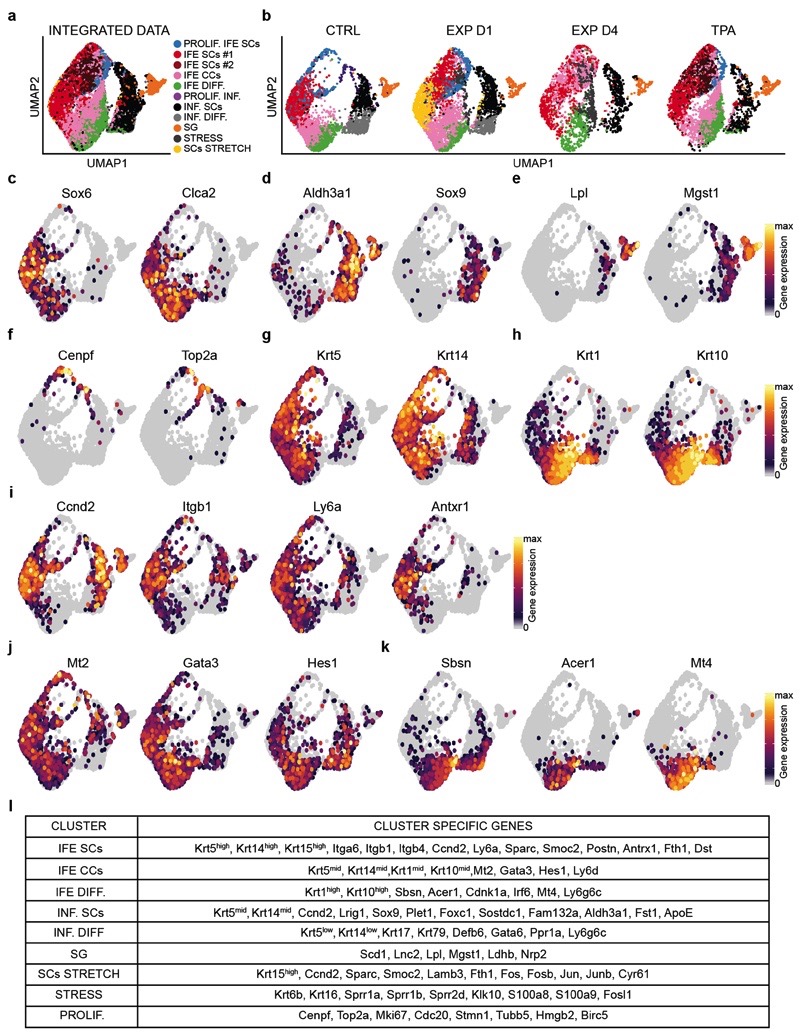
Extended Data Figure 6. Single-cell RNA sequencing clustering analysis.
a, Integrated Uniform Manifold Approximation and Projection (UMAP) graphic representation of the CTRL, EXP D1, EXP D4 and TPA single-cell RNA-seq data, showing the graph-based clustering results annotated by cell type. The proliferating IFE stem cells (PROLIF. IFE SCs) are in light blue, the IFE stem cells cluster are in red (IFE SCs#1) and dark red (IFE SCs#2), the IFE committed cells (IFE CCs) cluster is in pink and the differentiated IFE cells (IFE DIFF.) are in green. The differentiated cells from the infundibulum (INF. DIFF.) are in grey, the stem cells of the infundibulum (INF. SCs.) are in black, the proliferating cells of the infundibulum (PROLIF. INF.) are in plum and the sebaceous gland cluster (SG) is in orange. The IFE stress cells (STRESS) are in dark grey and the cluster of stem cells stretch (SCs STRETCH) in yellow. n=16651 cells. b, UMAP of the different samples (CTRL, EXP D1, EXP D4, TPA) using the same integrated projection. n=4659 cells CTRL, n= 4934 cells EXP D1, n= 2716 cells EXP D4, n= 4342 cells TPA. c-k, UMAP plot of the CTRL sample colored by normalized gene expression values for genes identifying the IFE (c) versus infundibulum (d), the sebaceous gland (e) and the proliferating cells (f). Undifferentiated (g) and more differentiated cells (h) in the IFE identified the SCs cluster (i), the CCs cluster (j) and the differentiated stage (k). Gene expression is visualized as a color gradient going from grey to yellow with grey as indicator of no expression (i.e. expression values below or equal to the 50th percentile for that sample) and yellow as indicator of maximal expression. c-k, n=16651 cells. l, Table showing the specific marker genes used to annotate the different clusters.