Abstract
Cancer development is frequently associated with dysregulation of mRNA translation to enhance both increased global protein synthesis and translation of specific mRNAs encoding oncoproteins. Thus, targeted inhibition of mRNA translation is viewed as a promising new approach for cancer therapy. In this article we review current progress in investigating dysregulation of mRNA translation initiation in mature B-cell neoplasms, focusing on chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma. We discuss mechanisms and regulation of mRNA translation, potential pathways by which genetic alterations and the tumor microenvironment alters mRNA translation in malignant B cells, preclinical evaluation of drugs targeted against specific eukaryotic initiation factors and current progress towards clinical development. Overall, inhibition of mRNA translation initiation factors is an exciting and promising area for development of novel targeted anti-tumor drugs.
Keywords: mRNA translation, initiation, lymphoma, leukemia, inhibitor, drug
Introduction
Although mRNA translation was viewed for many years as a constitutive, house-keeping function, it is now clear that this process is highly regulated and that its dysregulation can contribute to cancer development. Therefore, targeted inhibition of mRNA translation is a promising new approach to cancer therapy. In this review, we address the biological significance and drug targeting of mRNA translation pathways in mature B-cell neoplasms, a heterogeneous group of cancers that arise from cells of the adaptive immune system. We focus on pathways of B-cell lymphomagenesis/leukemogenesis, regulation of mRNA translation initiation and the effect on new drugs targeted against specific eukaryotic initiation factors (eIFs).
Mature B-cell neoplasms
Mature B-cell neoplasms arise from B cells which produce protective antibodies in response to foreign antigen. The cell surface B-cell receptor (BCR) plays a key role in determining the function of normal B cells since each clone within the B-cell repertoire has a unique BCR which defines the spectrum of antigens that bind [1]. This BCR diversity is generated by random recombination of immunoglobulin (Ig) gene segments followed by somatic hypermutation of the variable (antigen binding) regions. A diverse BCR repertoire is essential for health since it allows production of antibodies against a broad range of potential antigens. However, colateral effects of these mutagenic processes on non-Ig genes puts B cells at risk of accumulation additional mutations leading to activation of oncogenes/inactivation of tumor suppressors.
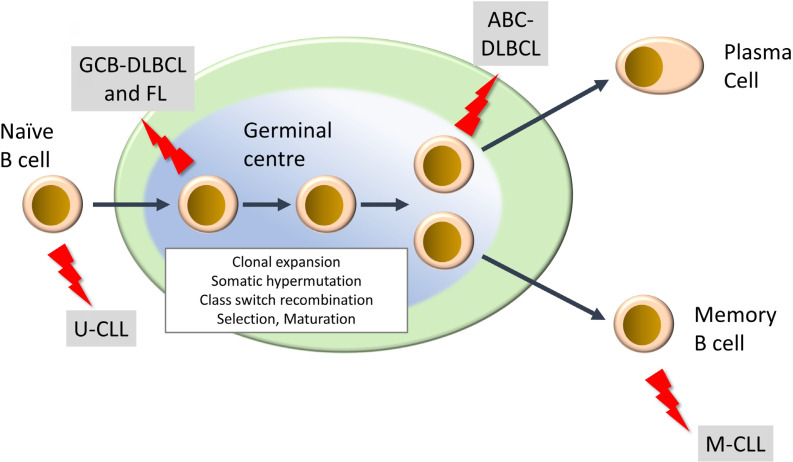
Mature lymphoid neoplasms are a strikingly diverse family of malignancies, both in terms of their etiology and clinical behavior, including response to treatment [2–4]. They are typically diagnosed in older individuals and their incidence is rising. There are more than 50 subtypes and this review will focus on the most common. This includes two subtypes of non-Hodgkin’s lymphoma, diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL), of which DLBCL is the most common aggressive B-cell lymphoma and FL is the most common indolent lymphoma. Chronic lymphocytic leukemia (CLL) is the most common B-cell leukemia. DLBCL and CLL can be further subdivided into activated B-cell-like (ABC) and germinal center B-cell-like (GCB) DLBCL, and unmutated CLL (U-CLL) and mutated CLL (M-CLL), respectively, and arise from different stages of B-cell differentiation within secondary lymphoid organs (Figure 1). In some cases, indolent disease can “transform” to a high-grade malignancy. In the UK, the annual incidence rates (per 100,000) for DLBCL, FL and CLL are 8.5, 3.4 and 7.2, respectively (https://www.hmrn.org/statistics/quick).
Figure 1.
Development of mature B-cell neoplasms. The germinal center (GC) is a key site for development of high affinity antibody responses. It is characterized by clonal expansion of B cells, somatic hypermutation (introduction of new mutations into Ig variable regions) and class switch recombination (where the constant region of the BCR is “switched” from IgM/IgD to other isotypes (IgG, IgA, IgE). There is also intense selection whereby B cells with higher affinity BCRs are positively selected whereas others are deleted by apoptosis. Selected B cells then mature to either antibody-producing plasma cells or memory B cells. “Lightning” symbols indicate transforming events leading to generation of FL and GCB-DLBCL from GC B cells and ABCDLBCL from a slightly more mature B cell termed a plasmablast. CLL comprises two main subsets derived from pre-GC (U-CLL) and post-GC (M-CLL) B cells
Due to the risk of off-target effects associated with Ig recombination, mature B-cell neoplasms are characterized by a high frequency of chromosomal translocations. For example, the t(14:18) translocation leads to BCL2 overexpression and suppression of apoptosis, and is a hallmark of FL [5]. The t(14:18) translocation is also found in GCB-DLBCL, but overall, the genetic complexity of DLBCL is greater than FL, and additional translocations may be present (e.g., leading to MYC overexpression) [6, 7]. There are also examples of “double hit” or “triple hit” DLBCL which are characterized by co-occurrence of mutations leading to activation of BCL2 with MYC and/or BCL6, and these are associated with a particularly poor outcome [8]. Translocations are much rarer in CLL and here BCL2 overexpression is commonly driven by loss of inhibitory miRNAs associated with recurrent deletion of part of chromosome 13 [9]. In addition to chromosomal changes, B-cell neoplasms are also characterized by somatic mutations affecting a wide range of pathways, including DNA damage (e.g., TP53, ATM), intracellular signaling pathways (e.g., MYD88, CD79B, NOTCH1, TNFRSF14), cell cycle control (CDKN2A) and epigenetics (e.g., EZH2, CREBBP) [2, 6, 7, 10, 11].
There is also an important role of the microenvironment in the development of these malignancies, including potential influences of T cells, stromal cells and cells of the innate immune system [12, 13]. Expression of the BCR is typically retained post-transformation and it is thought to play a major role by engaging pro-growth, proliferation and survival pathways, in response to antigen, environmental lectins or via autonomous BCR:BCR interactions [5, 14, 15].
Treatment for mature B-cell neoplasms typically relies on combinations of cytotoxics with anti-CD20 antibody therapy, for example, fludarabine, chlorambucil and rituximab (FCR) for CLL, and cyclophosphamide, doxorubicin, vincristine, prednisone and rituximab (CHOP-R) for FL/DLBCL [2, 3, 16]. DLBCL may be cured in 60–70% of cases but new treatments are required for a substantial proportion of patients, especially for ABCDLBCL and double/triple-hit DLBCL which have poorer outcomes. FL and CLL are both considered incurable although there may be a proportion of M-CLL cases with very long-term responses to FCR [17]. However, in most cases, initial responses can be promising but these low-grade malignancies typically follow a remitting/ relapsing course. New therapies targeted against cell survival pathways (e.g., venetoclax, a BCL2 inhibitor) or BCR-associated signaling kinases (e.g., ibrutinib and idelalisib, inhibitors of BTK and PI3K, respectively) can result in dramatic clinical responses and have revolutionized treatment in some cases [2, 18–21]. However, development of resistance and toxicity are major issues.
Overall, mature B-cell neoplasms are a heterogeneous group of malignancies which represent a significant and growing burden of morbidity and mortality. Existing treatments can be effective but there is a clear need for new treatment options, particularly for more aggressive disease subsets.
Mechanisms of mRNA translation
mRNA translation is the process by which the triplet code on mRNA is decoded by the ribosome to produce a polypeptide chain [22]. mRNA translation was considered for many years as a constitutive or “house-keeping” function. However, it is now clear that this process is highly regulated and that the mechanisms controlling mRNA translation are as sophisticated as those controlling transcription. Indeed, studies have indicated that modulation of mRNA translation may play as great a role as modulation of transcription in determining differences in protein expression between cell states [23]. Importantly, translational regulation provides a rapid mechanism to modulate protein expression compared to transcription. mRNA translation is usually considered in three steps, initiation, elongation and termination. mRNA translation initiation requires the action of multiple eIFs and accessory proteins, and is the most highly regulated step.
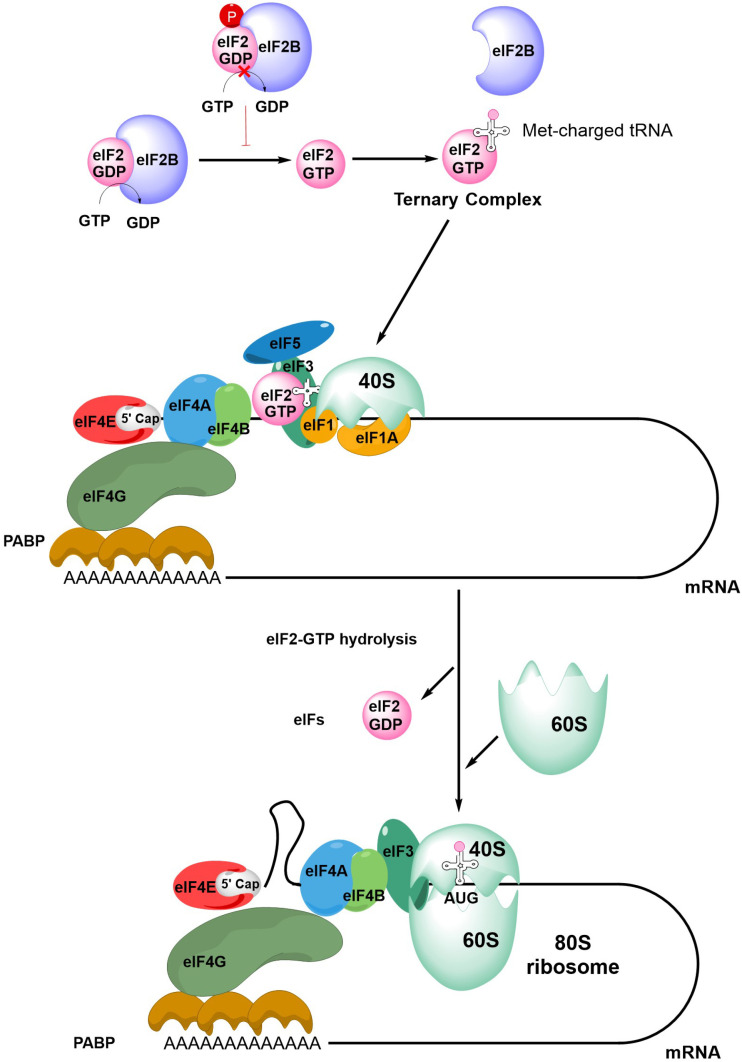
The first step in initiation of mRNA translation is formation of the eIF4F complex which contains eIF4A, eIF4E and eIF4G (Figure 2). eIF4E binds to the m7G-cap which is present at the 5′ end of mRNAs whereas eIF4A is a helicase which is important for unwinding mRNA secondary structures. eIF4G is a scaffold protein which binds eIF4A and eIF4E, and also contacts poly(A) binding protein (PABP) bound to the poly(A) 3’-tail of mRNA, thereby forming a circular messenger ribonucleoprotein complex, also known as the “closed-loop” structure. eIF4G/PABP interactions enhance mRNA translation initiation and can also influence recycling of ribosomes, translational termination and mRNA surveillance, for example, by effects on nonsense-mediated decay [24–27].
Figure 2.
An overview of mRNA translation initiation. The Figure shows some of the key steps in initiation of translation. The process initiates with the joining of the eIF4F complex (eIF4A, eIF4E and eIF4G), the tertiary complex, the 40S ribosome subunit and accessory eIFs. Interactions between eIF4G and PABP lead to closed-loop formation/mRNA circularization. The subsequent steps of scanning, initiation codon recognition and large ribosome subunit joining are accompanied by eIF2-GTP hydrolysis, release of eIF2-GDP and other eIFs. Note figure is illustrational and does not show all intermediate steps. See text for further details
Formation of the eIF4F:mRNA complex allows the recruitment of the ternary complex (TC) which contains GTP-bound eIF2 (a complex comprising α, β and γ subunits) and the methionine-loaded initiator tRNA. eIF2B is important for mRNA translation initiation since it is a guanine exchange factor required for GTP loading of eIF2α. The TC then binds the 40S ribosome subunit, an interaction that is stabilized by eIF1, eIF1A and eIF3, resulting in formation of the 43S pre-initiation complex (PIC) and binding of eIF5. This complex then “scans” the mRNA until it recognizes an initiation codon (usually AUG) in a suitable sequence context. Start site recognition is followed by arrest of the ribosome and hydrolysis of GTP within the TC. This triggers release of GDP-bound eIF2 and other eIFs and allows binding of the 60S ribosomal subunit to generate the 80S ribosome initiation complex.
Initiation is followed by elongation, whereby rounds of polypeptide bond formation add amino-acids to the nascent polypeptide chain. Finally, recognition of a termination codon leads to termination of mRNA translation and dissociation of the ribosome.
In addition to this cap-dependent mechanism of mRNA translation initiation, some mRNAs can be translated via alternate mechanisms [28]. For example, internal ribosome entry sites (IRES) in the 5′-untranslated region (UTR) of specific mRNAs can interact with canonical eIFs and IRES trans-acting factors to recruit ribosomes independently of the m7G-cap. Although originally described in viral mRNAs, IRESs facilitate translation of specific cellular mRNAs, especially in situations of stress, where m7G-cap-dependent mRNA translation is often inhibited. Of particular relevance to mature B-cell neoplasms, both the BCL2 and MYC oncoproteins can be translated via IRES-dependent mechanisms under at least some conditions [29, 30].
It is important to note that some eIFs have additional roles in mRNA-related processes. In particular, the m7G-cap binding function of eIF4E is exploited in the export of some mRNAs from the nucleus [31]. eIF4E’s role in RNA export is dependent on XPO1 (CRM1), which is itself required for export of multiple cargoes, including many proteins [32].
Selectivity and regulation of mRNA translation initiation
Variation in mRNA translation makes a major contribution to determining the differences in the proteome between cell types and cell states. Regulatory mechanisms influence overall translational capacity or can act on specific mRNAs to control expression of individual proteins. The expression of many proteins which are particularly relevant to biology of neoplastic B cells are subject to tight translational control, include regulators of cell growth (MYC), cell cycle progression (cyclin D1), and suppression of apoptosis (MCL1, BCL2). These are discussed in more detail below.
Some of the regulatory mechanisms that influence mRNA translation depend on intrinsic features present within the 5′-UTRs of target mRNAs that may be “sensed” by components of the eIF4F complex. For example, the helicase activity of eIF4A appears to be particularly important to unwind and facilitate translation of mRNAs with highly structured 5′-UTRs [33–36]. This includes mRNAs containing G-quadruplexes, RNA structures comprising stacked G-quartets stabilized by non-Watson-Crick interactions [33], which are predicted to occur in the 5′-UTRs of a substantial proportion of mRNAs, including BCL2 [37, 38]. These highly stable RNA structures are generally inhibitory for mRNA translation, either by preventing 43S PIC binding to mRNA or slowing scanning [39, 40].
Like eIF4A, eIF4E-dependency has also been linked to the presence of secondary structure but this may reflect the requirement of eIF4E for recruitment of eIF4A to the eIF4F complex. However, some specific mRNA 5′-UTR sequences have been linked to increased eIF4E-dependency. This includes so-called 5′-terminal oligopyrimidine (TOP) mRNAs which contain a cytidine residue at the m7G-cap followed by an uninterrupted sequence of pyrimidines [41]. Translation of these mRNAs is particularly sensitive to inhibition of mTORC1, a major upstream regulator of eIF4E activity [42, 43] (see below). However, not all mTORC1 sensitive mRNAs contain TOP sequences, and other specific sequence elements have also been linked to eIF4E dependency [43–45]. In mouse models, eIF4E haploinsufficiency is compatible with normal development but not for transformation, providing a potential explanation for the frequent activation of eIF4E in cancer cells [46]. Interestingly, mRNAs whose translation was particularly sensitive to eIF4E haploinsufficiency were frequently characterized by the presence of a 15 nucleotide 5′-UTR motif termed the cytosine enriched regulator of translation (CERT).
In addition to a requirement for the m7G-cap, dependency on eIF4E for mRNA nuclear export is also determined by specific mRNA sequences, specifically a ∼50-base 3’-UTR sequence element, known as 4E sensitivity element (4E-SE) [47]. mRNAs containing 4E-SE include CCND1, MYC and ODC1 and are particularly dependent on eIF4E for nuclear export.
Finally, it is important to note that several eIFs exist as multiple isoforms which may be functionally distinct. eIF4E2 was not thought to play a major role in mRNA translation initiation due its weaker m7G-cap and eIF4G binding activity, relative to eIF41. However, more recent data indicates that eIF4E2 may play a key role in sustaining mRNA translation in hypoxia [48]. Moreover, although there is considerable overlap in the mRNAs which are particularly sensitive to levels of eIF4E1 and eIF4E3, these factors are also associated with preferential translation of specific mRNAs via distinct 5′-UTR sequence motifs [49].
mRNA dependent translational control is also mediated by miRNAs which typically bind to sequences within the 3’-UTRs of target mRNAs [50]. Although miRNAs can induce degradation of their target mRNAs, their principle mode of action is via translational repression and some studies have indicated that the mechanism of miRNA-mediated translational repression is dependent on the eIF4A isoform eIF4A2 [51].
Various miRNAs have been linked to malignant features of B-cell neoplasms. As described above, deletion of part of chromosome 13 in CLL cells results in loss of miR-15a and miR-16-1, and subsequent suppression of apoptosis by increased BCL2 expression [9]. Other examples reveal the importance of complex networks linking B-cell neoplasm-associated proteins/pathways with multiple miRNAs [52]. For example, BCR signaling in CLL cells drives a malignancy promoting pathway mediated by increased expression of the miR-132-3p/miR-212 cluster, which in turn repress expression of TGFB1 and the negative cell cycle regulators EP400 and ZBTB5 [53]. In turn, BCR signaling is modulated by various miRNAs, including miR-155, miR150 and miR-17-92 [54–56]. There is also strong evidence that MYC is a central node in miRNA circuitry, since its expression is modulated by miRNAs (e.g., miR-34a and let-7) and it is itself a regulator of miRNA expression [57, 58].
mRNA translation is also controlled by multiple signaling inputs commonly activated downstream of cell surface receptors (Figure 3) [59]. These pathways can act globally to modulate overall mRNA translational capacity and/or influence translation of specific mRNAs to exert protein-specific control.
Figure 3.
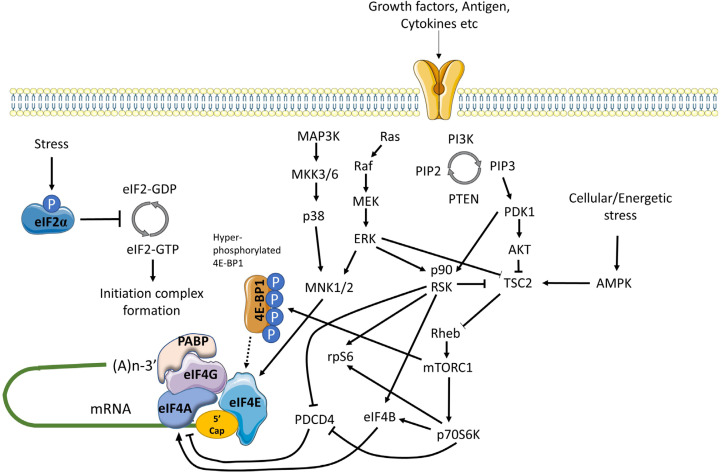
Positive and negative regulation of mRNA translation initiation by signaling pathways. mRNA translation is promoted by PI3K/AKT/mTORC1 and MAPK signaling. mTORC1 is activated via repression of TSC2-mediated conversion of Rheb-GTP to inactive Rheb-GDP. mTORC1 promotes the hyper-phosphorylation of 4E-BP1 which prevents inhibitory association with eIF4E, facilitating eIF4F complex formation. mTORC1 additionally activates p70S6K which indirectly activates eIF4A via repression of PDCD4 and activation of eIF4B. MAPK signaling leads to activation of MNK1/2 (and downstream phosphorylation of eIF4E) via MEK/ERK and p38 pathways, and ERK which with downstream-activated p90RSK, also relieves TSC2 mediated repression of mTORC1. In response to energetic stress AMPK is activated which promotes TSC2 mediated conversion of Rheb-GTP to Rheb-GDP to inhibit mTORC1. Other stress stimuli promote eIF2α phosphorylation that arrests the eIF2 complex in its inactive GDPbound form repressing mRNA translation
One of the key kinase complexes controlling mRNA translation is mTORC1 which comprises the mTOR kinase and various accessory proteins (raptor, GβL and deptor) [60]. The principal mTORC1 substrates for translational regulation include 4E-BP1 and p70S6K. Hypophosphorylated 4E-BPs interfere with eIF4F assembly by binding to a site on eIF4E that overlaps the eIF4E:eIF4G interaction interface. Thus, mTORC1-induced 4E-BP1 phosphorylation relieves inhibition of eIF4E and results in increased mRNA translation, including of TOP mRNAs [42, 43]. The function of p70S6K is less well understood, but its substrates include ribosomal protein S6 and eIF4B, an auxiliary factor that enhances eIF4A helicase activity. It can also phosphorylate and induce proteosomal degradation of PDCD4, an endogenous inhibitor of eIF4A [61].
mTORC1 activity is regulated by the TSC complex which inhibits mTORC1 activity by converting Rheb-GTP (an activator of mTORC1) into its inactive GDP-bound form [62]. Activation of AKT following receptor stimulation results in inhibitory phosphorylation of TSC2 (a component of the TSC complex), thereby relieving TSC-mediated repression of mTORC1 activity.
mRNA translation is also regulated by MAP kinase pathways, especially via ERK1/2 and p38 which can phosphorylate and activate MNK1/2 kinases [63]. Once activated, MNK1 phosphorylates eIF4E on Ser209 which appears to enhance its activity for both mRNA translation initiation and nuclear export. MNKs may also regulate mRNA translation initiation by phosphorylation of eIF4G. Interestingly, in DLBCL, MNK1 and MNK2 expression differs between the GCB and ABC subsets and this results in differential translation of a subset of mRNAs via effects on eIF4E1 and eIF4E3 isoforms [49].
Finally, there is cross-talk between the AKT/mTORC1 and MAPK pathways that influence mRNA translation. Perhaps most importantly, ERK1/2, and downstream p90RSK can promote inhibitory phosphorylation of TSC2, thereby contributing to activation of mTORC1 following receptor stimulation [59]. p90RSK can also phosphorylate rpS6 on Ser235/236 [59] and regulate eIF4A activity through phosphorylation of PDCD4 and eIF4B [64–66].
Cell stress can lead to a profound repression of mRNA translation, principally via an integrated stress response mediated by induced phosphorylation of eIF2α on Ser51 [67]. eIF2α Ser51 phosphorylation prevents guanine exchange by eIF2B, trapping eIF2 in its inactive GDP-bound form. eIF2α phosphorylation is catalyzed by one of four kinases PERK (activated following accumulation of unfolded proteins in the endoplasmic reticulum), PKR (activated in response to dsRNA during virus infection), HRI (activated by heme depletion, oxidative stress and heat-shock), and GCN2 (activated by amino-acid starvation). Whilst eIF2α phosphorylation is associated with profound repression of mRNA translation, translation of some mRNAs escapes this control allowing maintenance of protein expression (e.g., ATF4).
mRNA translation is extremely energy consuming and there are multiple mechanisms to ensure close coupling between mRNA translation capacity and ATP levels, and also with availability of amino-acids. Thus, amino-acid depletion negatively regulates mTORC1 activity via multiple mechanisms [68]. Moreover, a reduction in ATP availability leads to activation of AMPK which phosphorylates activatory sites on TSC2 leading to mTORC1 inactivation [60].
Finally, translational activity can be modulated by transcriptional control of its regulators. A key example is MYC which increases expression of mRNAs encoding eIF4A, eIF4E and eIF4G, as well as many other components involved in mRNA translation (e.g., ribosome components) [69, 70]. MYC mRNA translation is itself subject to tight translational control, suggesting the operation of a positive feedback loop. Another important factor that regulates mRNA translation via effects on transcription is the tumor suppressor p53 which represses expression of eIF4E [71].
Evidence linking alterations in mRNA translation initiation to mature B-cell neoplasms
Several studies have demonstrated that the expression of some eIFs/translational regulators differ between subtypes of mature B-cell neoplasms or between malignant B cells and their normal B-cell counterparts. Wang et al.[72], used immunohistochemistry of lymph node samples to demonstrate that expression of eIF4E and eIF2α was higher in more aggressive lymphoma subtypes compared to indolent subtypes. A subsequent study demonstrated that eIF4E expression in DLBCL was associated with poor outcome in both ABC and GCB subtypes [73]. Kodali et al.[74], demonstrated that 4E-BP1 expression was more commonly detectable in mature neoplasms (including FL, DLBCL and small lymphocytic lymphoma (SLL), the tissue-based counterpart of CLL) compared to non-malignant samples, whereas levels of 4E-BP1 phosphorylation were more variable between samples. Immunohistochemistry analysis also demonstrated that expression of the eIF4GII isoform was increased in DLBCL cells compared to normal B cells [75]. This may be linked to reduced expression of miR-520c-3p, a negative regulator of eIF4GII expression in DLBCL cells.
More recent studies using quantitative PCR demonstrated that 12/16 eIFs surveyed were overexpressed in DLBCL samples compared to normal controls [76]. Overexpression of eIF1A1, eIF3D and eIF2B5 was observed in both the GCB and non-GCB-DLBCL subtypes, but was somewhat greater in non-GCB-DLBCL, and was confirmed by immunohistochemistry. Interestingly, higher levels of eIF2B5, which is part of the eIF2B complex required for GTP loading of eIF2α, was associated with poor survival at both the RNA and protein level.
eIF4B (an activator of eIF4A) has been shown to be overexpressed in DLBCL cells compared to normal controls, and higher eIF4B expression was associated with poorer outcome [77]. This increased eIF4B expression was associated with enhanced translation of mRNAs encoding proteins involved in suppression of apoptosis and DNA repair, including BCL2, DAXX and ERCC5. One novel potential lymphoma-associated driver of eIF4B is fatty acid synthase (FASN) which stabilizes eIF4B and increase expression of oncoproteins such as MYC, BCL6 and MCL1 (a BCL2-related anti-apoptotic protein) via formation of a complex between eIF4B and the deubiquitinase USP11 in DLBCL cells [78].
Mouse models have also provided strong evidence that dysregulation of mRNA translation initiation contributes to lymphomagenesis. In particular, eIF4E co-operates with MYC to promote lymphomagenesis [79, 80]. Moreover, eIF4E overexpressing-lymphomas are relatively resistant to chemotherapy demonstrating that dysregulated mRNA translation can influence both tumor development and treatment responses [81]. More recent mechanistic analysis demonstrated that the ability of eIF4E to cooperate with MYC was dependent on the ability of eIF4E to bind the m7G-cap, but possibly not promote mRNA transport, at least in these models [82]. Interestingly, eIF4E activity was dependent on MNK1/2-mediated phosphorylation since a non-phosphorylatable mutant of eIF4E was inactive in these assays. Consistent with this, activated MNK1 also cooperated with MYC to promote lymphomagenesis. The anti-apoptotic MCL1 protein was identified as a key effector of this MNK/eIF4E pathway. Further linkage between mRNA translation initiation and lymphomagenesis is provided by observation that the deubiquitinase UCH-L1, which promotes assembly of eIF4F, is required for MYC-induced lymphomagenesis [83].
eIF6 plays a role in both ribosome biogenesis and mRNA translation initiation and seems to contribute to malignant transformation of B cells, since a reduction in eIF6 expression by deletion of single eIF6 allele is sufficient to reduce MYC-induced lymphomagenesis [84]. Interestingly, eIF6−/+ mice are healthy, indicating that malignant cells are particularly sensitive to the abundance of eIF6. eIF6 promotes glycolysis and fatty acid synthesis via increased translation of a subset of target mRNAs characterized by 5′-UTRs with a high G:C content or upstream open reading frames (both of which are generally inhibitory to translation) and reprogramming of metabolism by eIF6 may be required to support MYC-induced lymphomagenesis [84]. This critical dependency on eIF6 levels for transformation is reminiscent of eIF4E where haploinsufficiency is also sufficient to substantially reduce susceptibility to oncogene-induce transformation but is seemingly well tolerated by normal cells [46]. These are important observations since it suggests that may be possible to therapeutically inhibit eIF6 or eIF4E function in malignant cells with relatively little effect on normal cells.
Deletion of the eIF4A inhibitor PDCD4 has also been shown to increase lymphomagenesis in the mouse [85]. However, PDCD4 may have additional functions beyond translational control [86] and it is not clear to what extent increased eIF4A activity might contribute to increased lymphomagenesis in PDCD4-deficient animals.
Next generation sequencing has revealed the presence of recurrent mutations of genes encoding several factors related to mRNA translation, including XPO1, DDX3X, RPS15 and EIF2AK3 (PERK) in mature B-cell neoplasms [6, 7, 11]. Recurrent XPO1 mutations have been described in CLL and DLBCL, most commonly leading to amino-acid substitutions at E571. E571-mutant XPO1 co-operates with BCL2 or MYC to promote lymphomagenesis in vivo and is associated with altered nuclear/cytoplasmic distribution of various protein cargoes [87]. Mutant XPO1 also sensitized cells to the effects of the XPO1 inhibitor selinexor both in vitro and in vivo. However, the effects of XPO1 mutations on eIF4E-mediated mRNA transport have not been analyzed.
DDX3X is an RNA helicase that appears to play a specialized role in mRNA translation initiation, including non-AUG-initiated translation [88, 89]. Recurrent truncating mutations, leading to reduced DDX3X protein expression, have been described in CLL and DLBCL [90]. DDX3X mutations have also been described in medulloblastoma and more detailed functional analyses have shown that these mutations are associated with reduced global mRNA translation [91]. However, it is important to note that the spectrum of mutations of DDX3X differs between B-cell neoplasms and medulloblastoma (truncating mutations versus nonsynonymous single nucleotide changes) and the functional consequences of DLBCL/CLL-associated DDX3X mutations have not been reported.
Overall, several factors related to mRNA translation have been shown to be recurrently mutated in mature B-cell neoplasms and these may contribute to pathogenesis and influence drug responses. However, it is important to note that in the most common forms of mature B-cell neoplasms these mutations are found in a relatively small proportion (typically < 10%) of patients and most cases therefore do not appear to have mutations that directly dysregulate mRNA translation. However, there are some notable exceptions. For example, the frequency of XPO1 mutations may reach 50% in primary mediastinal B-cell lymphoma (a rare form of B-cell lymphoma) [92].
In the absence of mutations directly affecting the translational machinery, it seems likely that inappropriate mRNA translation in mature B-cell neoplasms is driven by upstream signaling pathways, activated by mutation or microenvironmental stimulation. Significant mutational drives likely include activation of mTORC1 signaling, e.g., due to loss of PTEN (a negative regulator of PI3K activity) in DLBCL or acquisition of mutations affecting RRAGC, which encodes an upstream activator of mTORC1, in FL [6, 7, 93]. Indeed, mTORC1 activity is required to drive the increased expression of eIF4B that is a feature of DLBCL [77]. In CLL, truncating mutations leading to activation of NOTCH1 signaling have been shown to be associated with increased BCR-induced mRNA translation [94], potentially mediated by a stimulatory effective of NOTCH1 signaling on the MNK/phospho-eIF4E pathway [95].
In terms of microenvironmental stimulation, it is likely that activation of the BCR plays a key role in driving mRNA translation in malignant B cells. Indeed, cross-linking of the BCR of CLL cells in vitro leads to increased global mRNA translation as well as translation of the MYC mRNA [96]. Moreover, gene expression analysis revealed that CLL cells derived from the lymph nodes of patients (the likely site of BCR stimulation of CLL cells by antigen in vivo) have increased expression of gene signatures related to mRNA translation compared to CLL cells in the blood. BCR-induced mRNA translation was associated with increased expression of eIF4A and eIF4G in CLL but not normal B cells, thereby revealing potential differences in mechanism of mRNA translational control between normal and malignant B cells.
In addition to eIFs, BCR signaling down-regulates expression of the eIF4A inhibitor PDCD4 [96, 97]. PDCD4 may be a common link between signaling and mRNA translational control since PDCD4 expression is downregulated following exposure of CLL cells to other microenvironmental signals, including the combination of bone marrow-derived stromal cells, CD40 ligand and CpG oligodeoxynucleotides (which stimulate toll-like receptor signaling), or the chemokine CXCL12 [98, 99].
CD40 ligand/IL4 signaling (to mimic T-cell help for B cells) has also been shown to increase eIF4E expression and 4EBP1 phosphorylation, and global mRNA translation in primary CLL cells [100]. Interestingly, this study also used ribosome profiling to show that several genes encoding proteins involved in the DNA damage response, including ATM, were particularly sensitive to translational control and that CD40 ligand/ IL4 treatment improved DNA damage responses in CLL cells exposed to X-radiation. Importantly, lower levels of ATM expression were associated with poorer outcome.
Finally, it is noteworthy that expression of many proteins with key roles in pathogenesis of mature B-cell neoplasms have been shown to be regulated at least in part at the level of mRNA translation (Table 1). Thus, dysregulated mRNA translation has potential to impinge widely of critical lymphoma/leukemiapromoting pathways.
Table 1.
Examples of translationally regulated genes with relevance to mature B-cell neoplasms
| Gene(s) | Protein function | Association with B-cell neoplasms | Evidence for translational control via eIFs | Refs |
|---|---|---|---|---|
| MYC | Cell growth |
|
|
[96, 101–104] |
| MCL1 | Cell survival (anti-apoptotic) |
|
|
[103] |
| BCL2 | Cell survival (anti-apoptotic) |
|
|
[101] |
| CCND1 | Cell cycle progression |
|
|
[103] |
| BCL6 | Transcriptional repression |
|
|
[101] |
| CARD11, BCL10, MALT1 | CBM complex components which control NF-κB activation |
|
|
[102] |
In vitro and in vivo effects of eIF inhibition
Consistent with the idea that dysregulated mRNA translation plays a key role in mature B-cell neoplasms, numerous preclinical studies have demonstrated anti-lymphoma/leukemia effects of inhibitors targeted against eIFs.
Analysis of eIF4A inhibitors has focused on naturally occurring flavaglines, particularly silvestrol which was originally isolated from the plant genus Aglaia. Silvestrol acts as a chemical inducer of dimerization by forcing interaction between free eIF4A (i.e., not complexed within eIF4F) and mRNA [105, 106] and preferentially reduces translation of mRNAs with highly structured 5′-UTRs [33, 34]. The closely structurallyrelated compound rocaglate A similarly “clamps” eIF4A to mRNAs, particularly those with polypurine tracts within their 5′-UTRs [107]. CRISPR/Cas9 editing has provided strong evidence that anti-cancer activities of flavaglines are mediated via eIF4A1 [108].
Using adoptive transfer of established Pten+/−Eμ-Myc and Eμ-Myc/eIF4E lymphoma cells, Bordeleau et al.[105], demonstrated that silvestrol was inactive when tested alone, but did enhance anti-lymphoma activity of doxorubicin. Interestingly, silvestrol did not show anti-lymphoma activity (alone or with doxorubicin) in an Eμ-Myc/Bcl2 model, suggesting that eIF4A inhibition specifically counters the in vivo growth of malignant B cells with a strong drive for translational dysregulation (i.e., either by activation of mTORC1 signaling downstream of PTEN loss or by enforced expression of eIF4E) but not suppression of apoptosis per se by BCL2. However, silvestrol alone has been shown to reduce lymphomagenesis in Eμ-Myc mice, where the latency of disease is relatively slow compared to adoptive transfer models [109].
Lucas et al.[110], investigated in vitro effects of silvestrol using primary CLL cells and demonstrated that silvestrol reduced expression of MCL1 protein and induced apoptosis of these cells (IC50 < 10 nM). Effects on MCL1 were independent of changes in MCL1 mRNA levels suggesting that silvestrol’s effects were mediated via translational inhibition. Silvestrol also improved survival in an Eμ-TCL1 mouse model, which recapitulates features of aggressive CLL.
Schatz et al.[103], demonstrated that silvestrol overcame protective effects associated with overexpression of the PIM2 kinase in an Eμ-Myc/Tsc2−/− in vivo lymphomagenesis model. Moreover, silvestrol induced apoptosis of various established B-lymphoma cell lines in vitro and down-modulated expression of cyclin D1, MYC and MCL1. Steinhardt et al.[102], demonstrated that silvestrol reduced expression of components of the CBM complex (an upstream regulator of NF-κB) in B-lymphoma cell lines following BCR stimulation whereas Wilmore et al.[104], showed that silvestrol reduced induction of MYC protein in primary CLL cells following BCR stimulation. Silvestrol has also been shown to induce apoptosis in various “MYC-driven” B-lymphoma cell lines [111] and this response was associated with reduced translation of MYC mRNA. This study also demonstrated that silvestrol synergized with the BCL2 inhibitor ABT-199 to kill lymphoma cell lines in vitro.
Cencic et al.[112], studied the alternate eIF4A inhibitor hippuristonal which was originally isolated from the coral Isis hippuris. Hippuristonal has a distinct mechanism of action compared to silvestrol and prevents both free and eIF4F-bound eIF4A from interacting with mRNA. Similar to silvestrol, hippuristonal had little activity alone in adoptive transfer models of MYC-induced lymphomagenesis, but cooperated with chemotherapeutics (doxorubicin or cyclophosphamide) to improve survival [113]. Interestingly, antilymphoma activity of hippuristonal was blocked by overexpression of the anti-apoptotic proteins BCL2 or MCL1 and hippuristonal synergized with the BCL2 inhibitor ABT-737 to induce of apoptosis of MYC lymphoma cells in vitro.
Other studies have focused on more recently identified eIF4A inhibitors, or on the development of synthetic analogues to overcome liabilities associated with natural product eIF4A inhibitors [111, 114, 115]. For example, silvestrol is a substrate for drug efflux pumps and this dramatically reduces oral bioavailability [116].
Chen et al.[117], recently reported synthetic derivatives of pateamine A which was originally isolated from the sea sponge Mycale sp. and like silvestrol, appears to promote association between eIF4A and mRNA. The derivative DMDAPatA induced apoptosis of CLL cells and synergized with ABT-199 to enhance cell killing. Some synthetic pateamine A analogues were shown to have reduced human plasma protein binding, a potential liability associated with the parental compound.
Peters et al.[118], used target-based screening approach to identify the natural product elatol (isolated from the red alga Laurencia microcladia) as an inhibitor of eIF4A1. In vitro analysis demonstrated that elatol reduced global mRNA translation and reduced expression of MYC, cyclin D3, MCL1, BCL2 and PIM2 in DLBCLderived cell lines. Like silvestrol, elatol also reduced induction of MYC in primary CLL cells following BCR stimulation and reduced the growth of B-lymphoma cells in vivo.
The pharmaceutical company eFFECTOR has reported a novel eIF4Ai (eFT226) which has shown promising in vitro and in vivo preclinical activity in models of lymphoma [119]. It has broad in vitro apoptosisinducing activity against cell lines derived from GCB-DLBCL, ABC-DLBCL and mantle cell lymphoma (a less common mature B-lymphoma) and reduces expression of MYC and MCL1. eFT226 also has strong in vivo activity against various B-lymphoma lines grown as xenografts in immunocompromised mice, even using a once-weekly dosing schedule.
The synthetic nucleoside ribavirin is used routinely as an anti-viral but has also received substantial attention as an eIF4E inhibitor. Ribavirin mimics the structure of the m7G-cap and competes with binding of the m7G-cap to eIF4E thereby inhibiting both mRNA translation initiation and nuclear mRNA export [31, 120].
Culjkovic-Kraljacic et al.[101], investigated the effect of ribavirin in models of double- or triple-hit DLBCL. They showed that ribavirin reduced the nuclear export and translation of BCL6, MYC and BCL2 mRNAs. Importantly, these effects were could be observed at concentrations of ribavirin (10 μM) that were similar to the steady state concentration of drug found in the plasma of patients undergoing anti-viral ribavirin treatment. Ribavirin reduced growth in vivo of a patient-derived xenograft derived from a triple hit DLBCL. Interestingly, ribavirin cooperated with an HSP90 inhibitor to suppress tumor growth (patient-derived lymphoma xenograft or xenografts of established double/triple-hit B-cell lines) potentially since HSP90 binds to eIF4E and may be required to maintain or enhance its activity. In CLL cells, ribavirin sensitizes to pro-apoptotic effects of fludarabine [121].
eFFECTOR has described a series of eIF4E inhibitors which compete for binding to the m7G-cap [122]. Ribosome profiling reveals that these compounds preferentially inhibit the translation of mRNAs containing TOP or CERT elements. Screening of a panel of human cell lines showed that cell lines derived from various hematological malignancies were particularly sensitive to growth inhibitory effects of these compounds, and that they also had substantial in vivo activity against the TMD8 (ABC-DLBCL-derived) cell line.
Small molecules have also been used to explore the effect of disruption of the eIF4:eIF4G interaction on malignant B cells. Cencic et al.[123], used molecular modelling and ultra-high throughput screening to identify 4E1RCat as an eIF4E:4G1 interaction inhibitor and showed that this compound effectively inhibited m7G-cap-dependent mRNA translation initiation and expression of MYC and MCL1 in Jurkat T-leukemia cells. 4EIRCat also cooperated with doxorubicin to extend survival in mice following adoptive transfer of Pten+/− Eμ-Myc lymphoma cells. Willimott et al.[124], demonstrated that a distinct eIF4E:4G1 interaction inhibitor, 4EGI-1, reduced translation of MCL1, BCL2A1 and BCL2L1 mRNAs, which encode anti-apoptotic BCL2-related proteins [125]. Moreover, 4EGI-1 and ABT-737 cooperated to promote CLL cell apoptosis.
Given the tight regulation of mRNA translation initiation by signaling, various kinase inhibitors have been used to examine the effects of indirect inhibition of eIFs by targeting of upstream activatory signaling. Most of this work has focused on inhibitors of mTORC1, especially rapamycin or related rapalogues (e.g., everolimus, temsirolimus). These compounds promote formation of a complex between mTORC1 and the immunophilin FKBP12 thereby reducing mTORC1 activity, whilst having relatively little effects on function of mTORC2 (an alternate mTOR kinase-containing complex), at least in short term experiments. Numerous studies have demonstrated ability of rapalogues to reduce proliferation or induce apoptosis of malignant B cells [126–129].
Despite this encouraging preclinical data, rapalogues have only been approved for treatment of mantle cell lymphoma and clinical responses in other B-cell neoplasms are generally modest [130–133]. The reasons for this modest clinical efficacy is not fully understand but may be due to incomplete inhibition of 4E-BP1 phosphorylation and/or stimulation of feedback loops leading to AKT activation which may be mediated via mTORC2 [134]. Thus, more recent efforts have focused on development of direct mTOR kinase inhibitors, which inhibit activity of both the mTORC1 and mTORC2 complexes, and dual mTOR/PI3K inhibitors [135, 136]. Such compounds have also demonstrated strong activity against malignant B cells in preclinical models, and in some cases have been shown to be superior to rapalogues [73, 137–139].
Given their important role in translational control, MNK inhibitors also offer considerable promise as potential therapies for B-cell neoplasms. eFFECTOR recently described the novel dual MNK1/2 inhibitor eFT508 and demonstrated that this compound inhibits the growth of a subset of DLBCL cell lines in vitro and has in vivo anti-lymphoma activity versus ABC-DLBCL xenografts [140, 141]. Consistent with MNK1/2 inhibition, eFT508 reduced eIF4E phosphorylation in vitro. Wu et al.[142], recently described QL-X-138, a dual inhibitor of MNKs and BTK, a pivotal kinase activated downstream of the BCR. QL-X-138 induced greater levels of apoptosis in primary CLL cells compared to ibrutinib or the MNK inhibitor cercosporamide alone.
Although not directly targeted towards eIFs, it is also worth considering inhibitors of XPO1 which is required for eIF4E to mediate its effects on nuclear export of mRNAs. Selective inhibitors of nuclear export (SINEs), including KPT-185, KPT-251 and selinexor, induce apoptosis of CLL cells in vitro, including samples with p53 dysfunction, and overcome the survival-promoting effects of microenvironmental stimuli [143]. Despite the importance of XPO1 in many cellular functions, normal B cells appear to be less sensitive to SINEinduced apoptosis compared to CLL cells. These compounds also have in vivo anti-leukemia activity in the Eμ-TCL1 model. Although SINEs have been shown to reduce protein nuclear export, effects on mRNA export and/or translation per se have not been investigated in these models. Selinexor also synergizes with ibrutinib to induce apoptosis of CLL cells in vitro and to improve anti-tumor activity in vivo in the Eμ-TCL1 model [144]. Moreover, selinexor retains activity in models of ibrutinib resistance.
Clinical observations
Clinical development of mRNA translation inhibitors has been led by the elongation inhibitor homoharringtonine (omacetaxine mepesuccinate) which is approved for third-line treatment of chronic myeloid leukemia [145]. Homoharringtonine activity is thought to be dependent on inhibition of translation of the BCR-ABL fusion protein which is critical for development of this leukemia.
The most advanced area of clinical testing related specifically to inhibition of mRNA translation initiation is for signaling inhibitors, especially targeted against mTORC1. As described above, promising preclinical data obtained with rapalogues has not resulted in substantial clinical progress, except for mantle cell lymphoma where temsirolimus is approved for treatment of relapsed/refractory disease. Moreover, the clinical benefit of next generation inhibitors, including dual mTORC1/2 and mTORC1/PI3K inhibitors, remains unclear [134, 146, 147]. Future clinical studies should focus on combinations to boost responses and identification of biomarkers which could reveal subset of responsive patients. For example, recent studies have shown that lymphoma-associated RRAGC mutations may confer enhanced sensitivity to mTORC1 inhibition [148].
The dual MNK1/2 inhibitor eFT508 is currently undergoing clinical evaluation as part of a phase 1/2 trial in patients with previously treated lymphoma (NCT02937675) [149].
The SINE selinexor has been approved for treatment of refractory multiple myeloma (derived from plasma cells) and has been evaluated for clinical activity in mature B-cell neoplasms. A phase 1 trial (NCT01607892) in patients with advanced hematological malignancies reported an overall objective response rate of 31%, although this was accompanied by significant toxicity [150]. On the basis of several complete responses, selinexor is currently being evaluated in a phase 2 trial in relapsed/refractory DLBCL (NCT02227251). Selinexor in combination with ibrutinib is also being evaluated in a phase 1 trial in patients with relapsed/ refractory CLL or aggressive NHL (NCT02303392). Other XPO1 inhibitors are in preclinical development and may offer a wider therapeutic window compared to selinexor [151].
Several case reports have described possible anti-lymphoma responses in individuals treated with ribavirin (alone or in combination with other anti-virals) for viral infection/reactivation [152, 153]. This includes a patient initially diagnosed with CLL that had undergone transformation to Hodgkin’s lymphoma who showed near complete response of lymph node lesions following treatment with ribavirin in the absence of any anti-lymphoma treatment [152]. It is notable that ribavirin has shown clinical activity in acute myeloid leukemia [154] and a trial to determine potential clinical activity of ribavirin in indolent FL and mantle cell lymphoma is underway (NCT03585725).
The eFFECTOR eIF4Ai (eFT226) is currently being evaluated for clinical activity in advanced solid tumor malignancies (NCT04092673) but will also be a very interesting candidate for clinical testing for hematological malignancies.
Conclusions
There has been a major shift in our perspective on mRNA translation, from a constitutive, house-keeping function to a highly regulated and selective process that is frequently dysregulated in cancer. In parallel, there has been a growing focus on targeted inhibition of mRNA translation as a novel strategy for cancer therapy. The field has progressed rapidly, and we now have much deeper insight into mechanisms of regulation and dysregulation of mRNA translation, and promising new chemical agents to target these pathways.
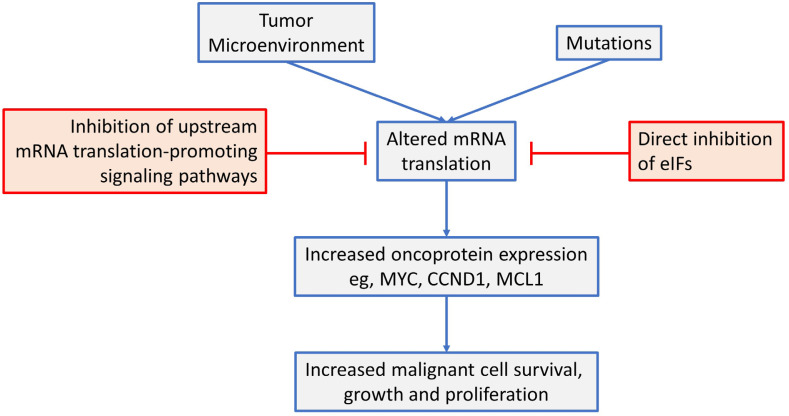
This review has focused on dysregulated ofk mRNA translation in common B-cell neoplasms (see Figure 4 for a summary). Overall, there is now strong evidence from mouse models that altered mRNA translation contributes to lymphomagenesis and that both gene mutations and signals from the tumor microenvironment are important drivers in human neoplasms. These alterations in mRNA translation initiation can lead to increased expression of critical oncoproteins that support the growth, proliferation and survival of malignant B cells, such as MYC, MCL1 and cyclin D1. Moreover, targeted inhibition of either upstream, mRNA translation promoting pathways (e.g., by inhibition of mTORC1 or MNKs), or direct inhibition of eIFs themselves can lead to dramatic anti-tumor effects in preclinical models, in vitro and in vivo. Importantly, mRNA translationtargeting agents can be combined effectively with chemotherapy and other targeted agents (including kinase inhibitors and BCL2 inhibitors) and this is likely to be critical to boost responses and reduce opportunities for development of resistance.
Figure 4.
An overview of mRNA translation initiation dysregulation in B-cell neoplasms and its targeted inhibition. See text for details
Clinical testing has been most advanced for drugs targeted against mTORC1, which is a powerful upstream activator of mRNA translation. However, despite impressive preclinical results, this has not resulted in substantial clinical advances. It will be particularly interesting to see emerging results obtained with newer agents targeted against other mRNA translation promoting pathways (e.g., MNK inhibitors) or directly against eIFs which are approaching or have entered early phase clinical trials.
New biological insight will be critical to underpin further development of mRNA translation inhibition as a novel therapeutic strategy. This should encompass analysis of upstream activating pathways which driver mRNA translation dysregulation and downstream consequences for expression of specific effector proteins and cellular pathways. This will be key to determine how best to target mRNA translation in different tumor types to maintain strong anti-cancer activity whilst minimizing toxicity. It will also identify molecular biomarkers that are required for both patient selection and pharmacodynamics monitoring of patient responses, and to guide the development of rationale drug combinations to boost responses and reduce development of resistance.
Abbreviations
- 4E-SE:
4E sensitivity element
- ABC:
activated B-cell-like
- BCR:
B-cell receptor
- CERT:
cytosine enriched regulator of translation
- CHOP-R:
cyclophosphamide, doxorubicin, vincristine, prednisone and rituximab
- CLL:
chronic lymphocytic leukemia
- DLBCL:
diffuse large B-cell lymphoma
- eIF:
eukaryotic initiation factor
- FASN:
fatty acid synthase
- FCR:
fludarabine, chlorambucil and rituximab
- FL:
follicular lymphoma
- GC:
germinal center
- GCB:
germinal center B-cell-like
- Ig:
immunoglobulin
- IRES:
internal ribosome entry site
- M-CLL:
mutated chronic lymphocytic leukemia
- PABP:
poly(A) binding protein
- PIC:
pre-initiation complex
- SINEs:
selective inhibitors of nuclear export
- SLL:
small lymphocytic lymphoma
- TC:
ternary complex
- TOP:
terminal oligopyrimidine
- U-CLL:
unmutated chronic lymphocytic leukemia
- UTR:
untranslated region
Declarations
Author contributions
All authors contributed to preparing the article, and read and approved the final submitted manuscript.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
The authors’ research on mRNA translation is funded by Cancer Research UK (C2750/A23669). The funder had no role in the preparation of this article.
Copyright
© The Author(s) 2020.
References
- 1.Monroe JG, Dorshkind K. Fate decisions regulating bone marrow and peripheral B lymphocyte development. Adv Immunol. 2007;95:1–50. 10.1016/S0065-2776(07)95001-4 [DOI] [PubMed] [Google Scholar]
- 2.Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3:16096. 10.1038/nrdp.2016.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017;390:298–310. 10.1016/S0140-6736(16)32407-2 [DOI] [PubMed] [Google Scholar]
- 4.Seifert M, Scholtysik R, Küppers R. Origin and pathogenesis of B cell lymphomas. Methods Mol Biol. 2019;1956:1–33. 10.1007/978-1-4939-9151-8_1 [DOI] [PubMed] [Google Scholar]
- 5.Küppers R, Stevenson FK. Critical influences on the pathogenesis of follicular lymphoma. Blood. 2018;131:2297–306. 10.1182/blood-2017-11-764365 [DOI] [PubMed] [Google Scholar]
- 6.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–90. 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–407. 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merron B, Davies A. Double hit lymphoma: how do we define it and how do we treat it? Best Pract Res Clin Haematol. 2018;31:233–40. 10.1016/j.beha.2018.07.012 [DOI] [PubMed] [Google Scholar]
- 9.Balatti V, Pekarky Y, Croce CM. Role of microRNA in chronic lymphocytic leukemia onset and progression. J Hematol Oncol. 2015;8:12. 10.1186/s13045-015-0112-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazarian G, Guièze R, Wu CJ. Clinical implications of novel genomic discoveries in chronic lymphocytic leukemia. J Clin Oncol. 2017;35:984–93. 10.1200/JCO.2016.71.0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krysiak K, Gomez F, White BS, Matlock M, Miller CA, Trani L, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017;129:473–83. 10.1182/blood-2016-07-729954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdière L, Mourcin F, Tarte K. Microenvironment signaling driving lymphomagenesis. Curr Opin Hematol. 2018;25:335–45. 10.1097/MOH.0000000000000440 [DOI] [PubMed] [Google Scholar]
- 13.Ten Hacken E, Burger JA. Microenvironment interactions and B-cell receptor signaling in chronic lymphocytic leukemia: implications for disease pathogenesis and treatment. Biochim Biophys Acta. 2016;1863:401–13. 10.1016/j.bbamcr.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packham G, Krysov S, Allen A, Savelyeva N, Steele AJ, Forconi F, et al. The outcome of B-cell receptor signaling in chronic lymphocytic leukemia: proliferation or anergy. Haematologica. 2014;99:1138–48. 10.3324/haematol.2013.098384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young RM, Phelan JD, Wilson WH, Staudt LM. Pathogenic B-cell receptor signaling in lymphoid malignancies: new insights to improve treatment. Immunol Rev. 2019;291:190–213. 10.1111/imr.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. 10.1182/blood-2016-01-643569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer K, Bahlo J, Fink AM, Goede V, Herling CD, Cramer P, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127:208–15. 10.1182/blood-2015-06-651125 [DOI] [PubMed] [Google Scholar]
- 18.Awan FT, Byrd JC. New strategies in chronic lymphocytic leukemia: shifting treatment paradigms. Clin Cancer Res. 2014;20:5869–74. 10.1158/1078-0432.CCR-14-1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerkeman M, Hallek M, Dreyling M, Thieblemont C, Kimby E, Staudt L. Targeting of B-cell receptor signalling in B-cell malignancies. J Intern Med. 2017;282:415–28. 10.1111/joim.12600 [DOI] [PubMed] [Google Scholar]
- 20.Okkenhaug K, Burger JA. PI3K signaling in normal B cells and chronic lymphocytic leukemia (CLL). Curr Top Microbiol Immunol. 2016;393:123–42. 10.1007/82_2015_484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yalniz FF, Wierda WG. Targeting BCL2 in chronic lymphocytic leukemia and other hematologic malignancies. Drugs. 2019;79:1287–304. 10.1007/s40265-019-01163-4 [DOI] [PubMed] [Google Scholar]
- 22.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 24.Fatscher T, Boehm V, Weiche B, Gehring NH. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA. 2014;20:1579–92. 10.1261/rna.044933.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. 10.1101/gad.1262905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. 10.1016/j.gene.2005.06.037 [DOI] [PubMed] [Google Scholar]
- 27.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson AG, Grosely R, Petrov AN, Puglisi JD. Dynamics of IRES-mediated translation. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160177. 10.1098/rstb.2016.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–74. 10.1074/jbc.M402727200 [DOI] [PubMed] [Google Scholar]
- 30.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–8. 10.1038/sj.onc.1201763 [DOI] [PubMed] [Google Scholar]
- 31.Osborne MJ, Borden KL. The eukaryotic translation initiation factor eIF4E in the nucleus: taking the road less traveled. Immunol Rev. 2015;263:210–23. 10.1111/imr.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickmanns A, Monecke T, Ficner R. Structural basis of targeting the exportin CRM1 in cancer. Cells. 2015;4:538–68. 10.3390/cells4030538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15:476. 10.1186/s13059-014-0476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–94. 10.1017/s135583820100108x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. 10.1038/cddis.2014.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahid R, Bugaut A, Balasubramanian S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry. 2010;49:8300–6. 10.1021/bi100957h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–8. 10.1093/nar/gkn511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19:158–74. 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Perreault JP, Topisirovic I, Richard S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin). 2016;4:e1244031. 10.1080/21690731.2016.1244031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849:801–11. 10.1016/j.bbagrm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- 42.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26:636–48. 10.1101/gr.197566.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita M, Gravel SP, Chénard V, Sikström K, Zheng L, Alain T, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 46.Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. 10.1016/j.cell.2015.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. 10.1083/jcb.200607020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uniacke J, Perera JK, Lachance G, Francisco CB, Lee S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer Res. 2014;74:1379–89. 10.1158/0008-5472.CAN-13-2278 [DOI] [PubMed] [Google Scholar]
- 49.Landon AL, Muniandy PA, Shetty AC, Lehrmann E, Volpon L, Houng S, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat Commun. 2014;5:5413. 10.1038/ncomms6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. 10.1038/cdd.2014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–5. 10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- 52.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29:1004–17. 10.1038/leu.2014.351 [DOI] [PubMed] [Google Scholar]
- 53.Pede V, Rombout A, Vermeire J, Naessens E, Mestdagh P, Robberecht N, et al. CLL cells respond to B-Cell receptor stimulation with a microRNA/mRNA signature associated with MYC activation and cell cycle progression. PLoS One. 2013;8:e60275. 10.1371/journal.pone.0060275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui B, Chen L, Zhang S, Mraz M, Fecteau JF, Yu J, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546–54. 10.1182/blood-2014-03-559690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K, et al. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood. 2014;124:84–95. 10.1182/blood-2013-09-527234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Psathas JN, Doonan PJ, Raman P, Freedman BD, Minn AJ, Thomas-Tikhonenko A. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood. 2013;122:4220–9. 10.1182/blood-2012-12-473090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao J, Zhao X, Tao J. c-MYC-miRNA circuitry: a central regulator of aggressive B-cell malignancies. Cell Cycle. 2014;13:191–8. 10.4161/cc.27646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol. 2012;4:a012252. 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. 10.1146/annurev-pharmtox-010611-134537 [DOI] [PubMed] [Google Scholar]
- 61.Carayol N, Katsoulidis E, Sassano A, Altman JK, Druker BJ, Platanias LC. Suppression of programmed cell death 4 (PDCD4) protein expression by BCR-ABL-regulated engagement of the mTOR/p70 S6 kinase pathway. J Biol Chem. 2008;283:8601–10. 10.1074/jbc.M707934200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8. 10.1038/embor.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joshi S, Platanias LC. Mnk kinase pathway: Cellular functions and biological outcomes. World J Biol Chem. 2014;5:321–33. 10.4331/wjbc.v5.i3.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galan JA, Geraghty KM, Lavoie G, Kanshin E, Tcherkezian J, Calabrese V, et al. Phosphoproteomic analysis identifies the tumor suppressor PDCD4 as a RSK substrate negatively regulated by 14-3-3. Proc Natl Acad Sci U S A. 2014;111:E2918–27. 10.1073/pnas.1405601111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–9. 10.1038/sj.emboj.7600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–91. 10.1038/sj.emboj.7601166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boye E, Grallert B. eIF2alpha phosphorylation and the regulation of translation. Curr Genet. 2019; [Epub ahead of print]. 10.1007/s00294-019-01026-1 [DOI] [PubMed] [Google Scholar]
- 68.Cao Y, Liu S, Liu K, Abbasi IHR, Cai C, Yao J. Molecular mechanisms relating to amino acid regulation of protein synthesis. Nutr Res Rev. 2019;32:183–91. 10.1017/S0954422419000052 [DOI] [PubMed] [Google Scholar]
- 69.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–34. 10.1158/0008-5472.CAN-07-5876 [DOI] [PubMed] [Google Scholar]
- 70.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–9. 10.1038/nrc2819 [DOI] [PubMed] [Google Scholar]
- 71.Zhu N, Gu L, Findley HW, Zhou M. Transcriptional repression of the eukaryotic initiation factor 4E gene by wild type p53. Biochem Biophys Res Commun. 2005;335:1272–9. 10.1016/j.bbrc.2005.08.026 [DOI] [PubMed] [Google Scholar]
- 72.Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen JJ, et al. Expression of the eukaryotic translation initiation factors 4E and 2alpha in non-Hodgkin’s lymphomas. Am J Pathol. 1999;155:247–55. 10.1016/s0002-9440(10)65118-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Demosthenous C, Han JJ, Stenson MJ, Maurer MJ, Wellik LE, Link B, et al. Translation initiation complex eIF4F is a therapeutic target for dual mTOR kinase inhibitors in non-Hodgkin lymphoma. Oncotarget. 2015;6:9488–501. 10.18632/oncotarget.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kodali D, Rawal A, Ninan MJ, Patel MR, Mesa H, Knapp D, et al. Expression and phosphorylation of eukaryotic translation initiation factor 4E binding protein 1 in B-cell lymphomas and reactive lymphoid tissues. Arch Pathol Lab Med. 2011;135:365–71. 10.1043/2009-0759-OA.1 [DOI] [PubMed] [Google Scholar]
- 75.Mazan-Mamczarz K, Zhao XF, Dai B, Steinhardt JJ, Peroutka RJ, Berk KL, et al. Down-regulation of eIF4GII by miR-520c-3p represses diffuse large B cell lymphoma development. PLoS Genet. 2014;10:e1004105. 10.1371/journal.pgen.1004105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Unterluggauer JJ, Prochazka K, Tomazic PV, Huber HJ, Seeboeck R, Fechter K, et al. Expression profile of translation initiation factor eIF2B5 in diffuse large B-cell lymphoma and its correlation to clinical outcome. Blood Cancer J. 2018;8:79. 10.1038/s41408-018-0112-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horvilleur E, Sbarrato T, Hill K, Spriggs RV, Screen M, Goodrem PJ, et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia. 2014;28:1092–102. 10.1038/leu.2013.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kapadia B, Nanaji NM, Bhalla K, Bhandary B, Lapidus R, Beheshti A, et al. Fatty acid synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat Commun. 2018;9:829. 10.1038/s41467-018-03028-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6. 10.1038/nm1042 [DOI] [PubMed] [Google Scholar]
- 80.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7. 10.1038/nature02369 [DOI] [PubMed] [Google Scholar]
- 81.Wendel HG, Lowe SW. Reversing drug resistance in vivo. Cell Cycle. 2004;3:847–9. [PubMed] [Google Scholar]
- 82.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–7. 10.1101/gad.1604407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hussain S, Bedekovics T, Liu Q, Hu W, Jeon H, Johnson SH, et al. UCH-L1 bypasses mTOR to promote protein biosynthesis and is required for MYC-driven lymphomagenesis in mice. Blood. 2018;132:2564–74. 10.1182/blood-2018-05-848515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miluzio A, Beugnet A, Grosso S, Brina D, Mancino M, Campaner S, et al. Impairment of cytoplasmic eIF6 activity restricts lymphomagenesis and tumor progression without affecting normal growth. Cancer Cell. 2011;19:765–75. 10.1016/j.ccr.2011.04.018 [DOI] [PubMed] [Google Scholar]
- 85.Hilliard A, Hilliard B, Zheng SJ, Sun H, Miwa T, Song W, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol. 2006;177:8095–102. 10.4049/jimmunol.177.11.8095 [DOI] [PubMed] [Google Scholar]
- 86.Matsuhashi S, Manirujjaman M, Hamajima H, Ozaki I. Control mechanisms of the tumor suppressor PDCD4: expression and functions. Int J Mol Sci. 2019;20:E2304. 10.3390/ijms20092304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor J, Sendino M, Gorelick AN, Pastore A, Chang MT, Penson AV, et al. Altered nuclear export signal recognition as a driver of oncogenesis. Cancer Discov. 2019;9:1452–67. 10.1158/2159-8290.CD-19-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phung B, Cieśla M, Sanna A, Guzzi N, Beneventi G, Cao Thi Ngoc P, et al. The X-linked DDX3X RNA helicase dictates translation reprogramming and metastasis in melanoma. Cell Rep. 2019;27:3573–86.e7. 10.1016/j.celrep.2019.05.069 [DOI] [PubMed] [Google Scholar]
- 89.Linsalata AE, He F, Malik AM, Glineburg MR, Green KM, Natla S, et al. DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep. 2019;20:e47498. 10.15252/embr.201847498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ojha J, Secreto CR, Rabe KG, Van Dyke DL, Kortum KM, Slager SL, et al. Identification of recurrent truncated DDX3X mutations in chronic lymphocytic leukaemia. Br J Haematol. 2015;169:445–8. 10.1111/bjh.13211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valentin-Vega YA, Wang YD, Parker M, Patmore DM, Kanagaraj A, Moore J, et al. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci Rep. 2016;6:25996. 10.1038/srep25996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jardin F, Pujals A, Pelletier L, Bohers E, Camus V, Mareschal S, et al. Recurrent mutations of the exportin 1 gene (XPO1) and their impact on selective inhibitor of nuclear export compounds sensitivity in primary mediastinal B-cell lymphoma. Am J Hematol. 2016;91:923–30. 10.1002/ajh.24451 [DOI] [PubMed] [Google Scholar]
- 93.Okosun J, Wolfson RL, Wang J, Araf S, Wilkins L, Castellano BM, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48:183–8. 10.1038/ng.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arruga F, Bracciamà V, Vitale N, Vaisitti T, Gizzi K, Yeomans A, et al. Bidirectional linkage between the B-cell receptor and NOTCH1 in chronic lymphocytic leukemia and in Richter’s syndrome: therapeutic implications. Leukemia. 2019;[Epub ahead of print]. 10.1038/s41375-019-0571-0 [DOI] [PubMed] [Google Scholar]
- 95.De Falco F, Sabatini R, Del Papa B, Falzetti F, Di Ianni M, Sportoletti P, et al. Notch signaling sustains the expression of Mcl-1 and the activity of eIF4E to promote cell survival in CLL. Oncotarget. 2015;6:16559–72. 10.18632/oncotarget.4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yeomans A, Thirdborough SM, Valle-Argos B, Linley A, Krysov S, Hidalgo MS, et al. Engagement of the B-cell receptor of chronic lymphocytic leukemia cells drives global and MYC-specific mRNA translation. Blood. 2016;127:449–57. 10.1182/blood-2015-07-660969 [DOI] [PubMed] [Google Scholar]
- 97.Perrot A, Pionneau C, Nadaud S, Davi F, Leblond V, Jacob F, et al. A unique proteomic profile on surface IgM ligation in unmutated chronic lymphocytic leukemia. Blood. 2011;118:e1–15. 10.1182/blood-2011-02-335125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carabia J, Carpio C, Abrisqueta P, Jiménez I, Purroy N, Calpe E, et al. Microenvironment regulates the expression of miR-21 and tumor suppressor genes PTEN, PIAS3 and PDCD4 through ZAP-70 in chronic lymphocytic leukemia. Sci Rep. 2017;7:12262. 10.1038/s41598-017-12135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O’Hayre M, Salanga CL, Kipps TJ, Messmer D, Dorrestein PC, Handel TM. Elucidating the CXCL12/CXCR4 signaling network in chronic lymphocytic leukemia through phosphoproteomics analysis. PLoS One. 2010;5:e11716. 10.1371/journal.pone.0011716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lezina L, Spriggs RV, Beck D, Jones C, Dudek KM, Bzura A, et al. CD40L/IL-4-stimulated CLL demonstrates variation in translational regulation of DNA damage response genes including ATM. Blood Adv. 2018;2:1869–81. 10.1182/bloodadvances.2017015560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Culjkovic-Kraljacic B, Fernando TM, Marullo R, Calvo-Vidal N, Verma A, Yang S, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood. 2016;127:858–68. 10.1182/blood-2015-05-645069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steinhardt JJ, Peroutka RJ, Mazan-Mamczarz K, Chen Q, Houng S, Robles C, et al. Inhibiting CARD11 translation during BCR activation by targeting the eIF4A RNA helicase. Blood. 2014;124:3758–67. 10.1182/blood-2014-07-589689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208:1799–807. 10.1084/jem.20110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilmore S, Yeomans A, Marriott S, Forconi F, Stevenson FK, Steele A, et al. Reversible inhibition of MYC translation in primary chronic lymphocytic leukaemia cells following eIF4A inhibition is associated with stabilization of MYC mRNA. Available from: https://abstracts.ncri.org.uk/abstract/reversible-inhibition-of-myc-translation-in-primary-chronic-lymphocytic-leukaemia-cells-following-eif4a-inhibition-is-associated-with-stabilization-of-myc-mrna/ [Last accessed on 01 Nov 2019].
- 105.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest. 2008;118:2651–60. 10.1172/JCI34753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cencic R, Carrier M, Galicia-Vázquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. 10.1371/journal.pone.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–61. 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chu J, Galicia-Vázquez G, Cencic R, Mills JR, Katigbak A, Porco JA, Jr, et al. CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep. 2016;15:2340–7. 10.1016/j.celrep.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lin CJ, Nasr Z, Premsrirut PK, Porco JA, Jr, Hippo Y, Lowe SW, et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep. 2012;1:325–33. 10.1016/j.celrep.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113:4656–66. 10.1182/blood-2008-09-175430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X, Bi C, Lu T, Zhang W, Yue T, Wang C, et al. Targeting translation initiation by synthetic rocaglates for treating MYC-driven lymphomas. Leukemia. 2020;34:138–150. 10.1038/s41375-019-0503-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–20. 10.1038/nchembio776 [DOI] [PubMed] [Google Scholar]
- 113.Cencic R, Robert F, Galicia-Vázquez G, Malina A, Ravindar K, Somaiah R, et al. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood Cancer J. 2013;3:e128. 10.1038/bcj.2013.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song J, Yuan C, Yang J, Liu T, Yao Y, Xiao X, et al. Novel flavagline-like compounds with potent Fli-1 inhibitory activity suppress diverse types of leukemia. FEBS J. 2018;285:4631–45. 10.1111/febs.14690 [DOI] [PubMed] [Google Scholar]
- 115.Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem. 2012;55:558–62. 10.1021/jm201263k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gupta SV, Sass EJ, Davis ME, Edwards RB, Lozanski G, Heerema NA, et al. Resistance to the translation initiation inhibitor silvestrol is mediated by ABCB1/P-glycoprotein overexpression in acute lymphoblastic leukemia cells. AAPS J. 2011;13:357–64. 10.1208/s12248-011-9276-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen R, Zhu M, Chaudhari RR, Robles O, Chen Y, Skillern W, et al. Creating novel translation inhibitors to target pro-survival proteins in chronic lymphocytic leukemia. Leukemia. 2019;33:1663–74. 10.1038/s41375-018-0364-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Peters TL, Tillotson J, Yeomans AM, Wilmore S, Lemm E, Jiménez-Romero C, et al. Target-based screening against eIF4A1 reveals the marine natural product elatol as a novel inhibitor of translation initiation with in vivo antitumor activity. Clin Cancer Res. 2018;24:4256–70. 10.1158/1078-0432.CCR-17-3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thompson PA, Eam B, Young NP, Fish S, Chen J, Barrera M, et al. Preclinical evaluation of eFT226, a novel, potent and selective eIF4A inhibitor with anti-tumor activity in B-cell malignancies. Blood. 2017;130:1530. 10.1182/blood.V130.Suppl_1.1530.1530 [DOI] [Google Scholar]
- 120.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–10. 10.1073/pnas.0406927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martinez-Marignac V, Shawi M, Pinedo-Carpio E, Wang X, Panasci L, Miller W, et al. Pharmacological targeting of eIF4E in primary CLL lymphocytes. Blood Cancer J. 2013;3:e146. 10.1038/bcj.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parker GS, Hung INJ, Staunton J, Barrera M, Sung E, Parra A, et al. Targeting PI3K/mTOR signaling with potent, selective and orally-available small molecule inhibitors of eIF4E. Available from: https://effector.com/wp-content/uploads/2018/12/eIF4E-AACR-PI3K-mTOR-2018-poster_final_for-distribution2.pdf. [Last accessed on 01 Nov 2019].
- 123.Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, et al. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci U S A. 2011;108:1046–51. 10.1073/pnas.1011477108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Willimott S, Beck D, Ahearne MJ, Adams VC, Wagner SD. Cap-translation inhibitor, 4EGI-1, restores sensitivity to ABT-737 apoptosis through cap-dependent and -independent mechanisms in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19:3212–23. 10.1158/1078-0432.CCR-12-2185 [DOI] [PubMed] [Google Scholar]
- 125.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–67. 10.1016/j.cell.2006.11.046 [DOI] [PubMed] [Google Scholar]
- 126.Decker T, Schneller F, Hipp S, Miething C, Jahn T, Duyster J, et al. Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin-dependent kinase (cdk) 4 and the cdk inhibitor p27. Leukemia. 2002;16:327–34. 10.1038/sj.leu.2402389 [DOI] [PubMed] [Google Scholar]
- 127.Wanner K, Hipp S, Oelsner M, Ringshausen I, Bogner C, Peschel C, et al. Mammalian target of rapamycin inhibition induces cell cycle arrest in diffuse large B cell lymphoma (DLBCL) cells and sensitises DLBCL cells to rituximab. Br J Haematol. 2006;134:475–84. 10.1111/j.1365-2141.2006.06210.x [DOI] [PubMed] [Google Scholar]
- 128.Muthukkumar S, Ramesh TM, Bondada S. Rapamycin, a potent immunosuppressive drug, causes programmed cell death in B lymphoma cells. Transplantation. 1995;60:264–70. 10.1097/00007890-199508000-00010 [DOI] [PubMed] [Google Scholar]
- 129.Witzig TE, Reeder C, Han JJ, LaPlant B, Stenson M, Tun HW, et al. The mTORC1 inhibitor everolimus has antitumor activity in vitro and produces tumor responses in patients with relapsed T-cell lymphoma. Blood. 2015;126:328–35. 10.1182/blood-2015-02-629543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bennani NN, LaPlant BR, Ansell SM, Habermann TM, Inwards DJ, Micallef IN, et al. Efficacy of the oral mTORC1 inhibitor everolimus in relapsed or refractory indolent lymphoma. Am J Hematol. 2017;92:448–53. 10.1002/ajh.24671 [DOI] [PubMed] [Google Scholar]
- 131.Witzig TE, Reeder CB, LaPlant BR, Gupta M, Johnston PB, Micallef IN, et al. A phase II trial of the oral mTOR inhibitor everolimus in relapsed aggressive lymphoma. Leukemia. 2011;25:341–7. 10.1038/leu.2010.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zent CS, LaPlant BR, Johnston PB, Call TG, Habermann TM, Micallef IN, et al. The treatment of recurrent/ refractory chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL) with everolimus results in clinical responses and mobilization of CLL cells into the circulation. Cancer. 2010;116:2201–7. 10.1002/cncr.25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Decker T, Sandherr M, Goetze K, Oelsner M, Ringshausen I, Peschel C. A pilot trial of the mTOR (mammalian target of rapamycin) inhibitor RAD001 in patients with advanced B-CLL. Ann Hematol. 2009;88:221–7. 10.1007/s00277-008-0582-9 [DOI] [PubMed] [Google Scholar]
- 134.Calimeri T, Ferreri AJM. m-TOR inhibitors and their potential role in haematological malignancies. Br J Haematol. 2017;177:684–702. 10.1111/bjh.14529 [DOI] [PubMed] [Google Scholar]
- 135.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–98. 10.1158/0008-5472.CAN-09-1751 [DOI] [PubMed] [Google Scholar]
- 136.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. 10.1158/0008-5472.CAN-05-2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Müller A, Zang C, Chumduri C, Dörken B, Daniel PT, Scholz CW. Concurrent inhibition of PI3K and mTORC1/mTORC2 overcomes resistance to rapamycin induced apoptosis by down-regulation of Mcl-1 in mantle cell lymphoma. Int J Cancer. 2013;133:1813–24. 10.1002/ijc.28206 [DOI] [PubMed] [Google Scholar]
- 138.Bi C, Zhang X, Lu T, Zhang X, Wang X, Meng B, et al. Inhibition of 4EBP phosphorylation mediates the cytotoxic effect of mechanistic target of rapamycin kinase inhibitors in aggressive B-cell lymphomas. Haematologica. 2017;102:755–64. 10.3324/haematol.2016.159160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yun S, Vincelette ND, Knorr KL, Almada LL, Schneider PA, Peterson KL, et al. 4EBP1/c-MYC/PUMA and NF-kappaB/EGR1/BIM pathways underlie cytotoxicity of mTOR dual inhibitors in malignant lymphoid cells. Blood. 2016;127:2711–22. 10.1182/blood-2015-02-629485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reich SH, Sprengeler PA, Chiang GG, Appleman JR, Chen J, Clarine J, et al. Structure-based design of pyridone-aminal eFT508 targeting dysregulated translation by selective mitogen-activated protein kinase interacting kinases 1 and 2 (MNK1/2) inhibition. J Med Chem. 2018;61:3516–40. 10.1021/acs.jmedchem.7b01795 [DOI] [PubMed] [Google Scholar]
- 141.Webster KR, Goel VK, Hung IN, Parker GS, Staunton J, Neal M, et al. eFT508, a potent and selective mitogen-activated protein kinase interacting kinase (MNK) 1 and 2 inhibitor is efficacious in preclinical models of diffuse large B-cell lymphoma (DLBCL). Blood. 2015;126:1554. 10.1182/blood.V126.23.1554.1554 [DOI] [Google Scholar]
- 142.Wu H, Hu C, Wang A, Weisberg EL, Chen Y, Yun CH, et al. Discovery of a BTK/MNK dual inhibitor for lymphoma and leukemia. Leukemia. 2016;30:173–81. 10.1038/leu.2015.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhong Y, El-Gamal D, Dubovsky JA, Beckwith KA, Harrington BK, Williams KE, et al. Selinexor suppresses downstream effectors of B-cell activation, proliferation and migration in chronic lymphocytic leukemia cells. Leukemia. 2014;28:1158–63. 10.1038/leu.2014.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hing ZA, Mantel R, Beckwith KA, Guinn D, Williams E, Smith LL, et al. Selinexor is effective in acquired resistance to ibrutinib and synergizes with ibrutinib in chronic lymphocytic leukemia. Blood. 2015;125:3128–32. 10.1182/blood-2015-01-621391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.LüS, Wang J. Homoharringtonine and omacetaxine for myeloid hematological malignancies. J Hematol Oncol. 2014;7:2. 10.1186/1756-8722-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brown JR, Hamadani M, Hayslip J, Janssens A, Wagner-Johnston N, Ottmann O, et al. Voxtalisib (XL765) in patients with relapsed or refractory non-Hodgkin lymphoma or chronic lymphocytic leukaemia: an open-label, phase 2 trial. Lancet Haematol. 2018;5:e170–80. 10.1016/S2352-3026(18)30030-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ghobrial IM, Siegel DS, Vij R, Berdeja JG, Richardson PG, Neuwirth R, et al. TAK-228 (formerly MLN0128), an investigational oral dual TORC1/2 inhibitor: a phase I dose escalation study in patients with relapsed or refractory multiple myeloma, non-Hodgkin lymphoma, or Waldenström’s macroglobulinemia. Am J Hematol. 2016;91:400–5. 10.1002/ajh.24300 [DOI] [PubMed] [Google Scholar]
- 148.Ortega-Molina A, Deleyto-Seldas N, Carreras J, Sanz A, Lebrero-Fernández C, Menéndez C, et al. Oncogenic Rag GTPase signaling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Nat Metab. 2019;1:775–89. 10.1038/s42255-019-0098-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gopal AK, Ramchandren R, Gabrail N, Patel MR, Wang D, Miller LL, et al. A phase 1–2 dose-escalation and cohort-expansion study of eFT508, a selective, orally bioavailable inhibitor of MNK1 and MNK2, in patients with hematological malignancies. Blood. 2017;130 Suppl 1:S4624. 10.1182/blood.V130.Suppl_1.4624.4624 [DOI] [Google Scholar]
- 150.Kuruvilla J, Savona M, Baz R, Mau-Sorensen PM, Gabrail N, Garzon R, et al. Selective inhibition of nuclear export with selinexor in patients with non-Hodgkin lymphoma. Blood. 2017;129:3175–83. 10.1182/blood-2016-11-750174 [DOI] [PubMed] [Google Scholar]
- 151.Hing ZA, Fung HY, Ranganathan P, Mitchell S, El-Gamal D, Woyach JA, et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia. 2016;30:2364–72. 10.1038/leu.2016.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rutherford SC, Stewart EN, Chen Z, Chadburn A, Wehrli NE, van Besien K, et al. The eIF4E inhibitor ribavirin as a potential antilymphoma therapeutic: early clinical data. Leuk Lymphoma. 2018;59:256–8. 10.1080/10428194.2017.1323270 [DOI] [PubMed] [Google Scholar]
- 153.Maciocia N, O’Brien A, Ardeshna K. Remission of follicular lymphoma after treatment for hepatitis C virus infection. N Engl J Med. 2016;375:1699–701. 10.1056/nejmc1513288 [DOI] [PubMed] [Google Scholar]
- 154.Assouline S, Culjkovic-Kraljacic B, Bergeron J, Caplan S, Cocolakis E, Lambert C, et al. A phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia with elevated eIF4E. Haematologica. 2015;100:e7–9. 10.3324/haematol.2014.111245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.