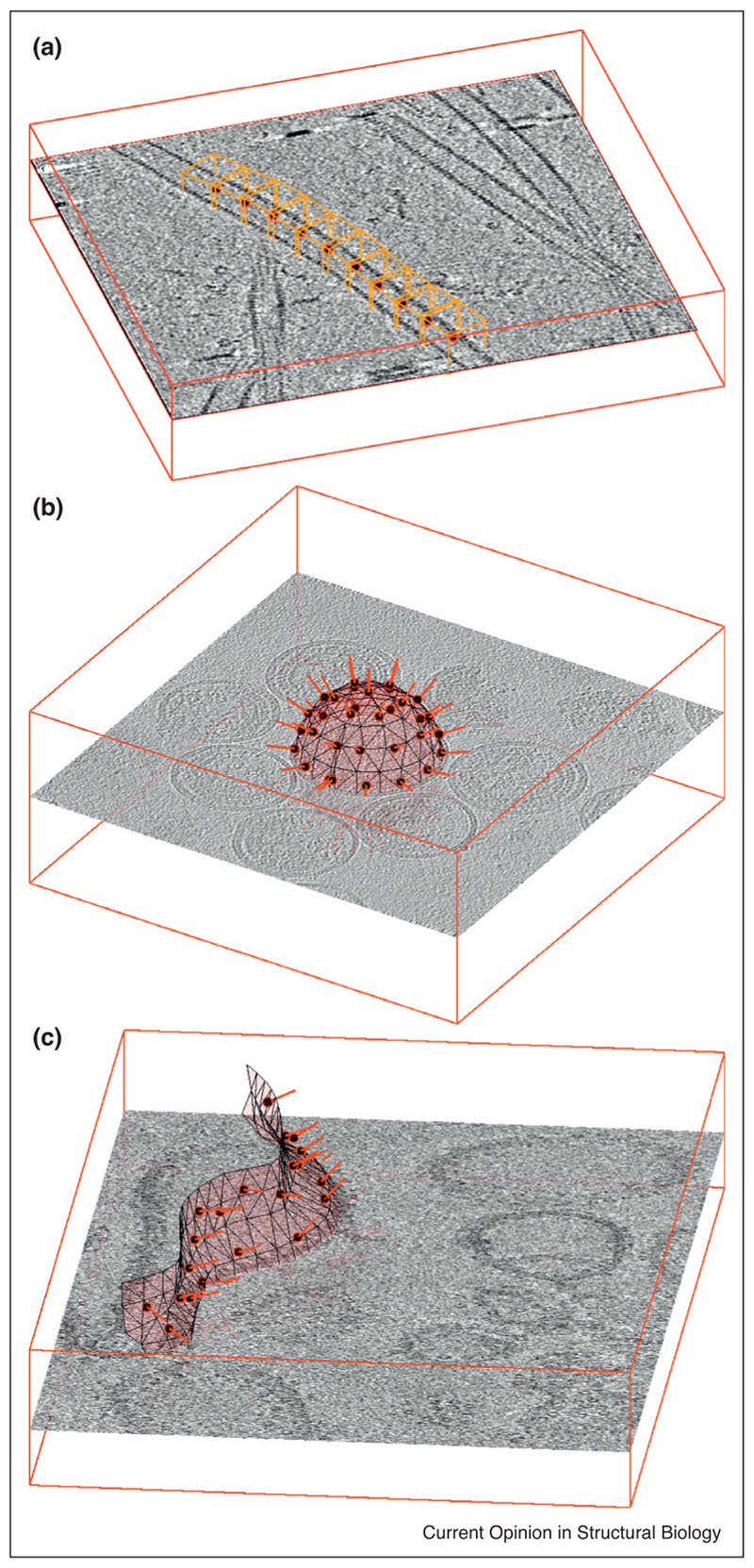
Figure 2.
Examples of particle extraction for (a) filaments, (b) vesicles and (c) generically shaped membranes. In all three cases, the position of a subtomogram (red circle filled in black) is accompanied by an initial estimation of the orientation of the particle (red bar) given by the respective support geometry. In (a), orange boxes representing subtomograms are placed along the axis of a microtubule (data of microtubule-bound dynein-dynactin complexes from Grotjahn et al. [25]. The initial orientation of each subtomogram is given by the local orientation of the microtubule. In (b) the particles of interest are GAG proteins forming the capsid of HIV virus-like particles (VLPs) in solution (data from EMPIAR-10164). Their extraction is facilitated by using estimations of radius and centres to model the surface of the VLP as a sphere: locations of subtomograms are regularly distributed on it, while the local normals provide initial orientations. The tomogram in (c) shows membrane-assembled, in vitro reconstituted murine 5HT3 serotonins. The membrane of a non-spherical liposome is modelled as a triangulated surface, allowing the placement of a regular distribution of subtomogram centres and initial orientation assignations.