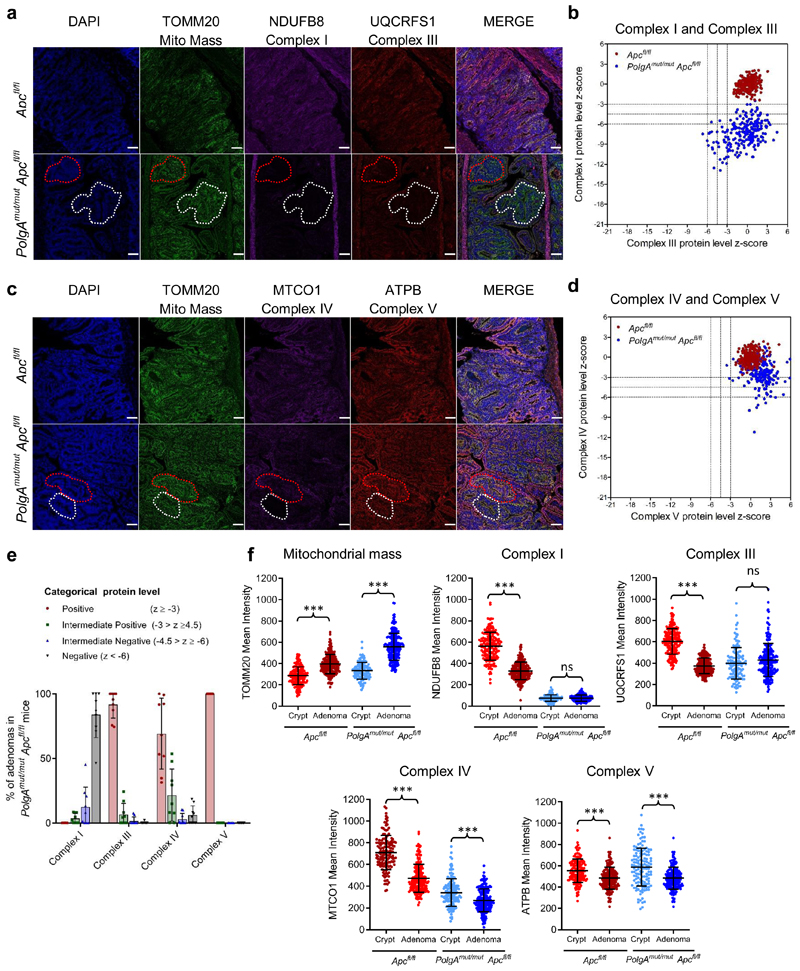
Figure 4. Small Intestinal adenomas from PolgAmut/mut;Apcfl/fl mice are deficient in mitochondrial Complex I, but the majority retain expression of subunits of complexes III, IV and V.
a and c: Immunofluorescence was performed to quantify levels of OXPHOS proteins on n=9 PolgAmut/mut;Apcfl/fl mice and n=10 Apcfl/fl mice. Representative images are shown. Scale bars 50µm. The white dashed line shows an adenoma region deficient in complex I and III, and the red dashed line highlights deficiency in complex I only in panel a: An adenoma region deficient in complex IV is highlighted by the white dashed line and one with normal complex IV is highlighted by the red dashed line in panel c. b and d: Dot plots showing relative Z-scores calculated following quantification of mitochondrial OXPHOS protein levels in adenomas from PolgAmut/mut;Apcfl/fl (n=9) and Apcfl/fl (n=10) mice using the method in29. n=20 adenomas were quantified per mouse. e: Categorical analysis of OXPHOS protein levels PolgAmut/mut;Apcfl/fl (n=9 mice) and Apcfl/fl (n=10 mice) Data points show individual mice ± s.d. f: Dot plots showing raw densitometry values for mitochondrial protein levels. For the adenomas: n=9 PolgAmut/mut;Apcfl/fl and n=10 Apcfl/fl mice with 20 adenomas analysed per mouse. For the normal crypts, n=5 mice were analysed per group with a minimum of 13 crypts quantified per mouse. One-way ANOVA with Tukey’s post-test. P values for within genotype comparisons between normal crypts and adenomas were as follows: TOMM20: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p<0.0001, NDUFB8: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.9995, UQCRFS1: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.1302, MTCO1: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.0001, ATPB: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p<0.0001. For all panels: * p<0.05, **p<0.01, ***p<0.001, error bars show s.e.m