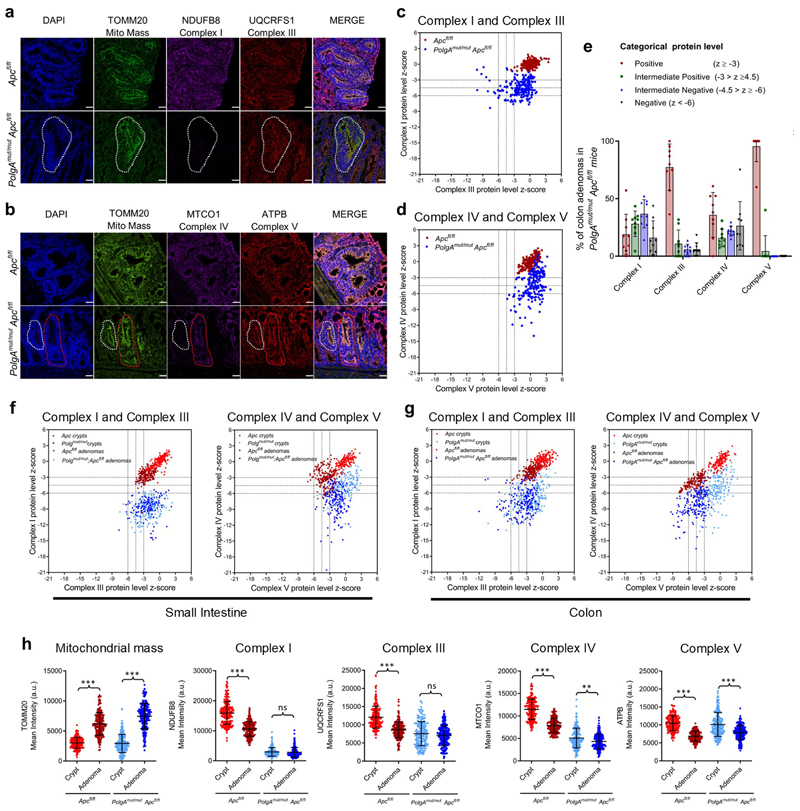
Extended Data Fig. 2. Colonic adenomas from PolgAmut/mut;Apcfl/fl mice are deficient in mitochondrial complex I, but the majority retain expression of subunits of complexes III, IV and V.
a and b: Immunofluorescence was performed to quantify levels of OXPHOS proteins in n=9 PolgAmut/mut;Apcfl/fl mice and n=9 Apcfl/fl mice. Representative images are shown. Scale bars 50µm. An adenoma deficient in complex I is highlighted by the white dashed line in a. The white dashed line highlights an adenoma deficient in complex IV, and red dashed line shows one with normal complex IV in b. c and d: dot plots showing Z-scores calculated following quantification of mitochondrial OXPHOS protein levels in adenomas from n=9 PolgAmut/mut;Apcfl/fl and n=9 Apcfl/fl mice with 20 adenomas quantified per mouse. e : Categorical analysis of OXPHOS protein levels in PolgAmut/mut;Apcfl/fl (n=9) and Apcfl/fl (n=9) mice, error bars show mean ±s.d. f-g : dot plots showing Z-scores calculated following quantification of mitochondrial OXPHOS protein levels in normal crypts and adenomas in the small intestine (f) and the colon (g). f : For the adenomas: n=9 PolgAmut/mut;Apcfl/fl and n=10 Apcfl/fl mice were analysed with 20 adenomas quantified per mouse. For the normal crypts, n=5 mice were analysed with a minimum of 13 crypts quantified per mouse. g : For the colonic adenomas: n=9 mice per group were analysed with a minimum of 20 adenomas quantified per mouse. For the normal crypts, n=6 Apcfl/fl mice and n=7 PolgAmut/mut;Apcfl/fl mice were analysed with a minimum of 22 crypts quantified per mouse. h Dot plots showing raw densitometry values for mitochondrial protein levels in the colon (n numbers same as in g, error bars are s.d.). One-way ANOVA with Tukey’s post-test. P values for within genotype comparisons between normal crypts and adenomas were as follows: TOMM20: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p<0.0001, NDUFB8: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.9761, UQCRFS1: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.2901, MTCO1: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p=0.0007, ATPB: Apcfl/fl p<0.0001, PolgAmut/mut;Apcfl/fl p<0.0001. For all panels: * p<0.05, **p<0.01, ***p<0.001