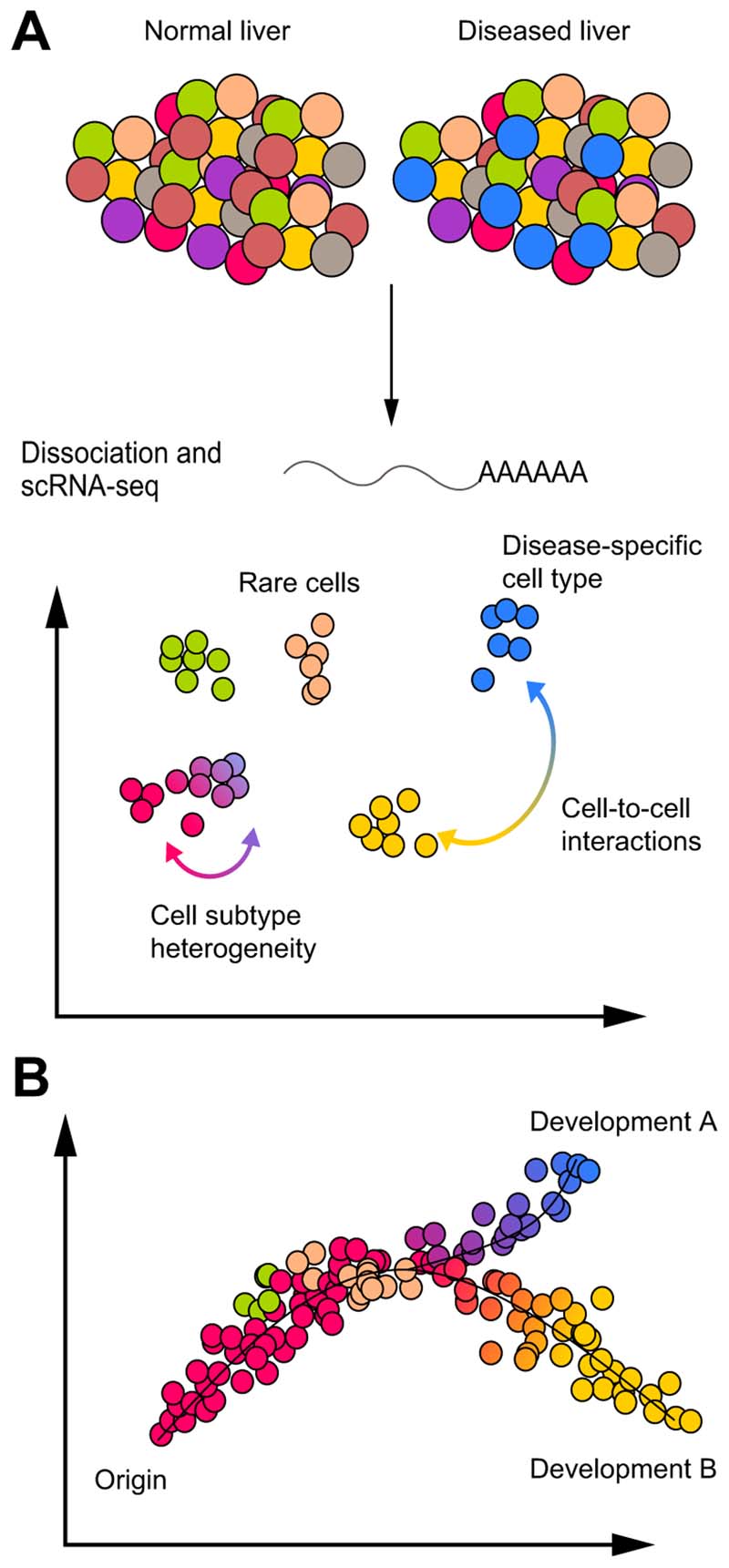
Fig. 1. Single-cell RNA-sequencing analyses to study liver pathophysiology.
(A) Normal and/or diseased liver tissue are dissociated into a single cell suspension and scRNA-seq is performed. Thousands of transcripts per cell are compressed in a 2D space where each cell is a dot and the distance between cells is a function of their similarity. Cells are can be aggregated in clusters or groups of clusters plotted as different colours and potentially representing cell types or subtypes. ScRNA-seq allows the study of rare cell types, cell state and subtype heterogeneity, disease-specific cell type and cell-to-cell interactions via ligand-receptor analysis. (B) Computational analyses such as pseudo-time diffusion mapping or RNA velocity, which analyse cell similarity and diversity, consent to trace differentiation processes, clonal evolution and cell state transitions of a specific cell type or between different cell types (from cell of origin to development A or B). ScRNA-seq, single-cell RNA-sequencing.