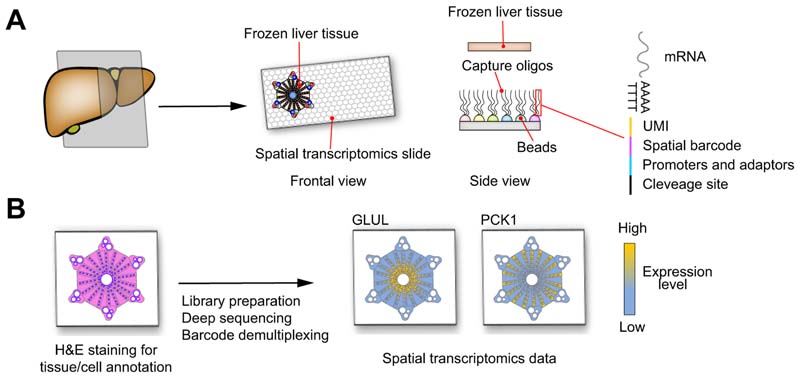
Fig. 4. Model of in situ spatial transcriptomics.
(A) Frozen liver tissue is cut and placed on a special slide special slide covered by beads carrying capture oligos composed by a polyd(T) tail for RNAs capture, a spatial barcode defining bead position, an UMI for transcript count, promoters and adaptors for cDNA synthesis, amplification and sequencing and a cleavage site to detach the oligos from the slide. B) The liver tissue on the spatial transcriptomics slide is fixed, stained by H&E and scanned by a conventional microscopy slide scanner. The tissue is lysed to release RNA, the capture oligos are cleaved and the libraries prepared as for scRNA-seq. The H&E image combined with data and coordinates of the spatial barcodes produce high-resolution single-cell gene expression data. GLUL encoding for glutamate-ammonia ligase is a known pericentral zonated gene and PCK1 encoding for phosphoenolpyruvate carboxykinase 1 is a periportal zonated gene. cDNA, complementary DNA; scRNA-seq, single-cell RNA-sequencing; UMI, unique molecule identifier.