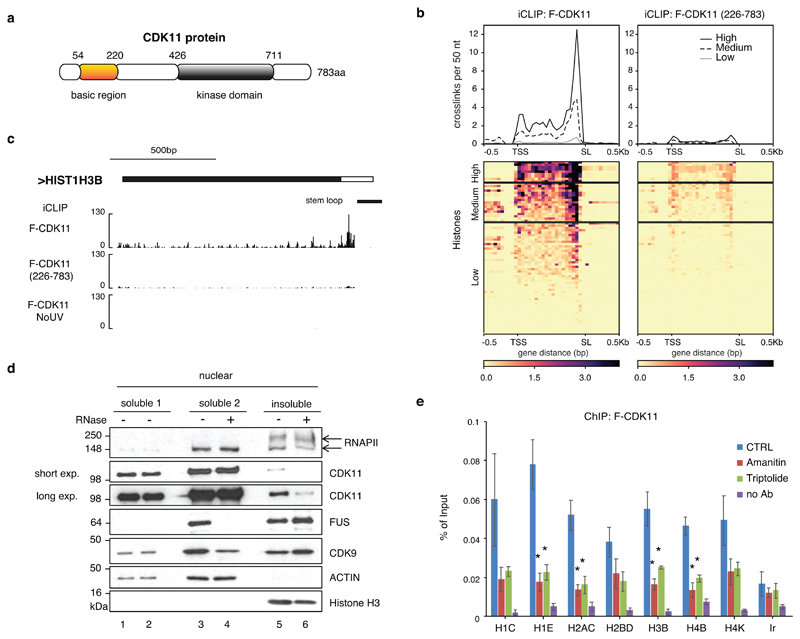
Figure 4. RNA promotes CDK11 recruitment to the RDH chromatin.
a, Schematic diagram highlighting the kinase domain and basic region of human CDK11 protein.
b, Metagene analyses of F-CDK11 and F-CDK11 (226-783) iCLIP binding at all RDH transcripts from the TSS to the SL. iCLIP data k-means clustered, based on RNA-seq expression (high, medium and low). n=4 biologically independent experiments.
c, Biodalliance genome browser view of F-CDK11, F-CDK11 (226-783) and uncrosslinked control (no UV) iCLIP binding at HIST1H3B transcript. Stem loop (SL) is indicated by a black line.
d, Western blot analysis of association of the indicated factors in soluble and insoluble fractions of chromatin either treated or not treated with RNase A/T1. Arrows mark phosphorylated (upper) and non-phosphorylated (lower) forms of RNAPII. For CDK11, long and short exposures of the film are shown.
e, CDK11 ChIP-qPCR on RDH genes in HCT116 cells expressing stably integrated F-CDK11 and treated either with Amanitin (4 μg/ml) or Triptolide (10 μM) or untreated (CTRL). n=4 biologically independent experiments, error bars=SEM, *P<0.05, Student´s t-test, Ir=intergenic region. Source data for panel e are available with the paper on line.