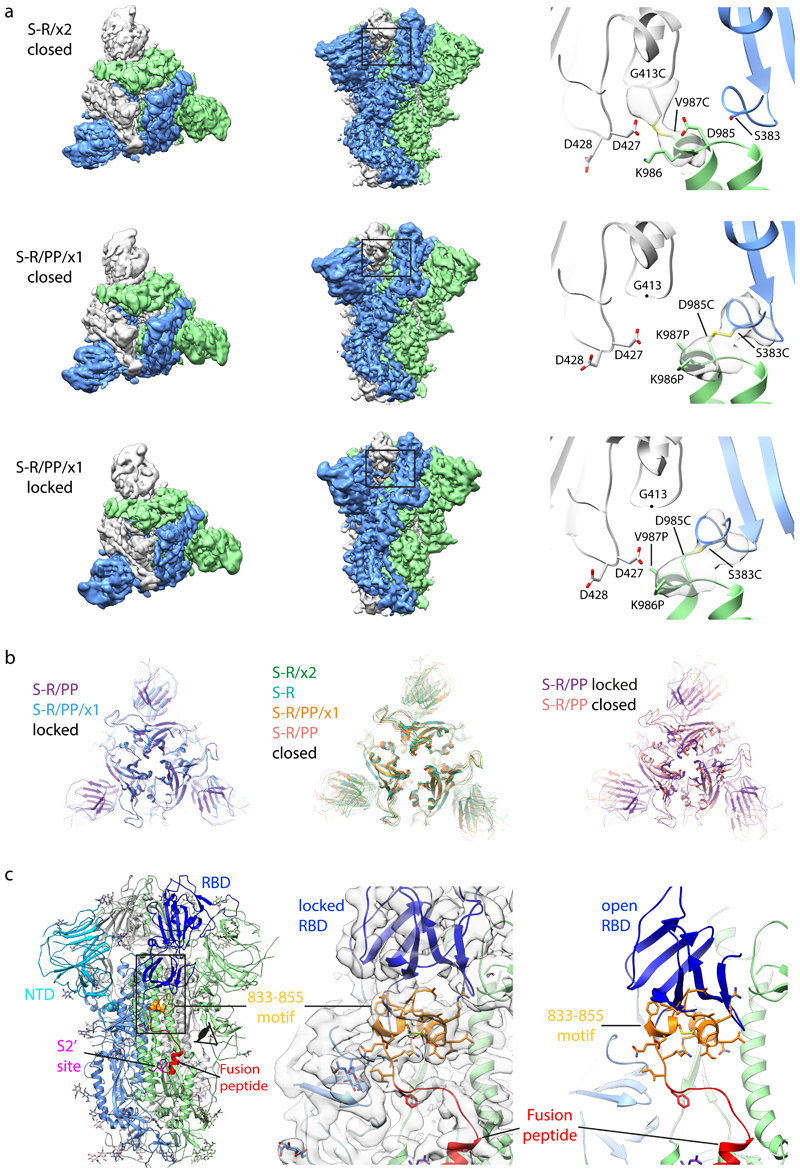
Figure 3. Characterisation of S-protein trimers by cryo-EM.
a) Cryo-EM densities for S-R/x2 in the closed conformation, and for S-R/PP/x1 in both closed and locked conformations. Boxes mark regions shown in right hand panels which illustrate resolved density for the inserted disulphide bonds at the expected positions. b) Comparison of the RBD and NTD conformations in S trimers of locked and closed conformations as viewed from the top of the trimer at the three-fold axis after alignment based on the central S2 helices. Between the locked and closed conformations the S1 domains show a twist motion (right panel and compare left and middle panel). In the locked conformation, the RBD moves closer to the 3-fold axis. The position of the NTD is variable. c) Folding of 833-855 into a helix-turn-helix motif locks the RBD in a locked conformation. Left panel, locations of RBD, 833-855 motif (orange), fusion peptide and S2' site are coloured and indicated. Middle panel, detailed density for the 833-855 motif is viewed from near the fusion peptide towards the RBD (indicated by the eye icon in the left panel). The density shows well resolved density that was disordered in previous SARS-CoV-1 and SARS-CoV-2 S structures. Right panel, the position of the RBD in the open conformation is aligned onto the locked monomer. In the open conformation the RBD would clash into the folded 833-855 motif (orange).