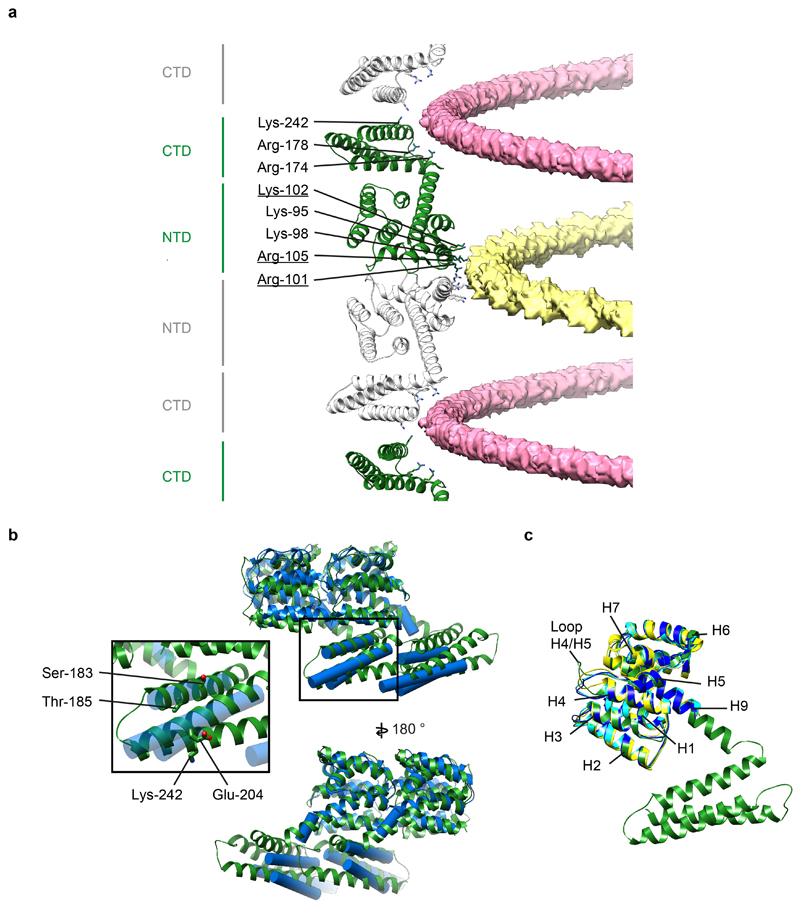
Extended Data Figure 6. Analysis of the in vitro M1 structure.
a) A view of M1 monomers extracted from the helical reconstruction to show the sites of interaction with the two nucleic acid strands (see also Fig. 2). One nucleic acid strand binds at the NTD-NTD interface (yellow), the other binds to a groove formed at the CTD-CTD interface (pink). Residues interacting with nucleic acid are all positively charged and are shown as sticks. Residues that form part of the nuclear localisation signal which was previously shown to bind the viral ribonucleoprotein 64 are underlined. b) Alignment of in situ (blue) and in vitro (green) M1 dimers extracted from their respective linear polymers. The differences are limited to small movements at the interfaces, perturbations in the orientation of α helix 9 that accommodate the different curvatures, and a small change in the orientation of α helix 12. It is possible that these differences reflect differences between spherical PR8 virions and filamentous HK68 virions, but we think it is more likely that they reflect the different curvature and the presence of nucleic acid in the in vitro sample. Inset highlights selected residues in the CTD. The mutations Ser183Ala and Thr185Ala cause spherical IAV WSN to make more filamentous particles 65. We speculate that these mutations may modulate folding of the CTD. Residue 204 is Glu in filamentous Udorn and HK68, but is Asp in spherical WSN, and the mutation Glu204Asp reduces the number of long filaments 21. This residue is close to the C-terminal end of the neighbouring CTD and we speculate that this difference may influence this interaction. Residue 242 can be sumoylated 66. This residue faces the inside of the virion where sumoylation could be accommodated without altering M1 packing. c) Alignment of the full-length M1 structure determined by helical reconstruction (green) to crystal structures of M1 NTD – PDB:1AA7 (blue) 12, PDB:1EA3 (yellow) 13, and PDB:5V6G (cyan) 14. The structures are the same except for small differences in the H4/H5 loop.