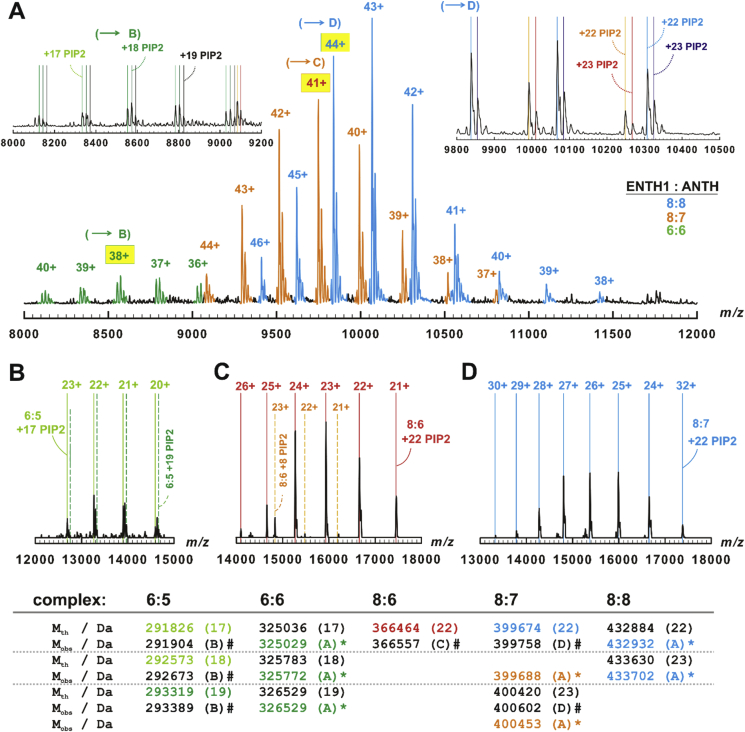
Fig. 1.
Higher order complex formation of yeast ENTH1 and ANTH. A: Orbitrap mass spectrum of yeast ENTH1 and ANTH assembling into di-hexamers, pentadecamers (8:7) and di-octamers (green, orange and cyan peaks, respectively). Capital letters and hatched-in charge labels denote the peaks selected for MS/MS (B-D). Insets: Lower (left) and upper (right) parts of the spectrum with theoretical m/z of the di-hexamer (green) as well as of the pentadecamer (8:7) (orange, red) and the di-octamer (cyan, blue) with 17–19, 22 or 23 PIP2 ligands indicated by vertical lines. B-D: Relevant parts of fragment ion mass spectra of the di-hexamer (B), the pentadecamer (8:7) (C), and of the di-octamer (D) complexes. Vertical lines denote the theoretical m/z of hexa-pentameric (B), of octa-hexameric (C), and pentadecameric (8:7) stripped (D) complexes with the indicated number of PIP2 ligands. Lower panel: comparison of theoretical (Mth) and experimentally determined (Mobs) masses. The font color mirrors measured (A) or theoretical (B-D) m/z; numbers in brackets indicate the amount of PIP2 ligands. #: determined by MS/MS. *: determined from two independent precursor ion mass spectra, difference was always below 0.5 Da.