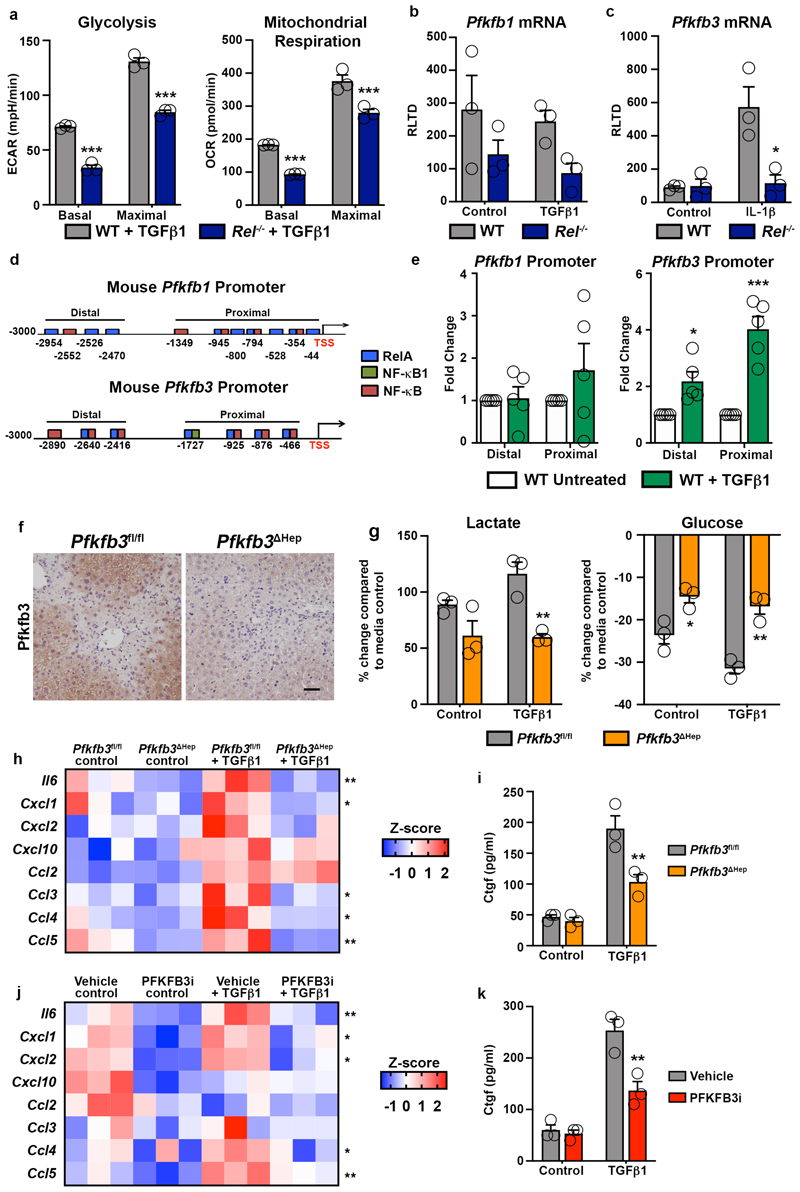
Extended Data Fig. 3. c-Rel regulates metabolic enzymes to induce epithelial dedifferentiation and fibrogenic gene expression.
(a) Seahorse analysis of basal (p<0.0001) and maximal (p<0.0001) glycolysis (extracellular acidification rate, ECAR) and basal (p<0.0007) and maximal (p<0.0004) mitochondrial respiration (oxygen consumption rate, OCR) in WT and Rel-/- hepatocytes stimulated ± TGFβ1. (b) Graph shows mRNA expression of Pfkfb1 in WT and Rel-/- hepatocytes stimulated ± TGFβ1. (c) Graph shows mRNA expression of Pfkfb3 in WT and Rel-/- hepatocytes stimulated ± IL-1β. (p value=0.025) (d) Schematic representation of RelA, NF-κB1 and NF-κB binding sites in the murine Pfkfb1 and Pfkfb3 promoters. (e) ChIP analysis of c-Rel at the proximal and distal regions of the Pfkfb1 promoter and the proximal (p<0.0001) and distal (p=0.0185) regions of the Pfkfb3 promoter in WT hepatocytes treated ± TGFβ1. (f) Representative images show PFKFB3 immunohistochemical staining in liver sections from acute CCl4 injured Pfkfb3flfl and Pfkfb3 Δhep mice. Images are representative of n=5 mice/group. Scale bar is 100(m. (g) Graphs show media lactate in control and TGFβ1 (p=0.0064) stimulated and glucose levels in control (p=0.0227) and TGFβ1 (p=0.00284) stimulated in hepatocytes isolated from Pfkfb3flfl and Pfkfb3 Δhep mice and stimulated ± TGFβ1. (h) Heatmap showing secreated Il-6, Cxcl1, Cxcl2, Cxcl10, Ccl2, Ccl3, CCl4 and Ccl5, measured by MSD in the media of hepatocytes isolated from Pfkfb3flfl and Pfkfb3Δhep mice and stimulated ± TGFβ1. (i) Quantification of connective tissue growth factor (CTGF) in pg/ml in the culture media of hepatocytes isolated from Pfkfb3flfl and Pfkfb3Δhep mice and stimulated ± TGFβ1 (p=0.0027) (j) Heatmap showing secreated Il-6, Cxcl1, Cxcl2, Cxcl10, Ccl2, Ccl3, CCl4 and Ccl5, measured by MSD in the media of WT hepatocytes stimulated ± TGFβ1 ± the Pfkfb3 inhibitor 3PO. (k) Quantification of connective tissue growth factor (CTGF) in pg/ml in the culture media of WT hepatocytes stimulated ± TGFβ1 ± the Pfkfb3 inhibitor 3PO (p=0.0013). Data in graphs are mean ± s.e.m. in n=3 (g,i,k), n=4 (a,b,c) or n=5 (e) independent cell isolations/condition. All p values were calculated using a two-way ANOVA with Tukey post-hoc t-test (* P <0.05, ** P <0.01 and *** P <0.001).