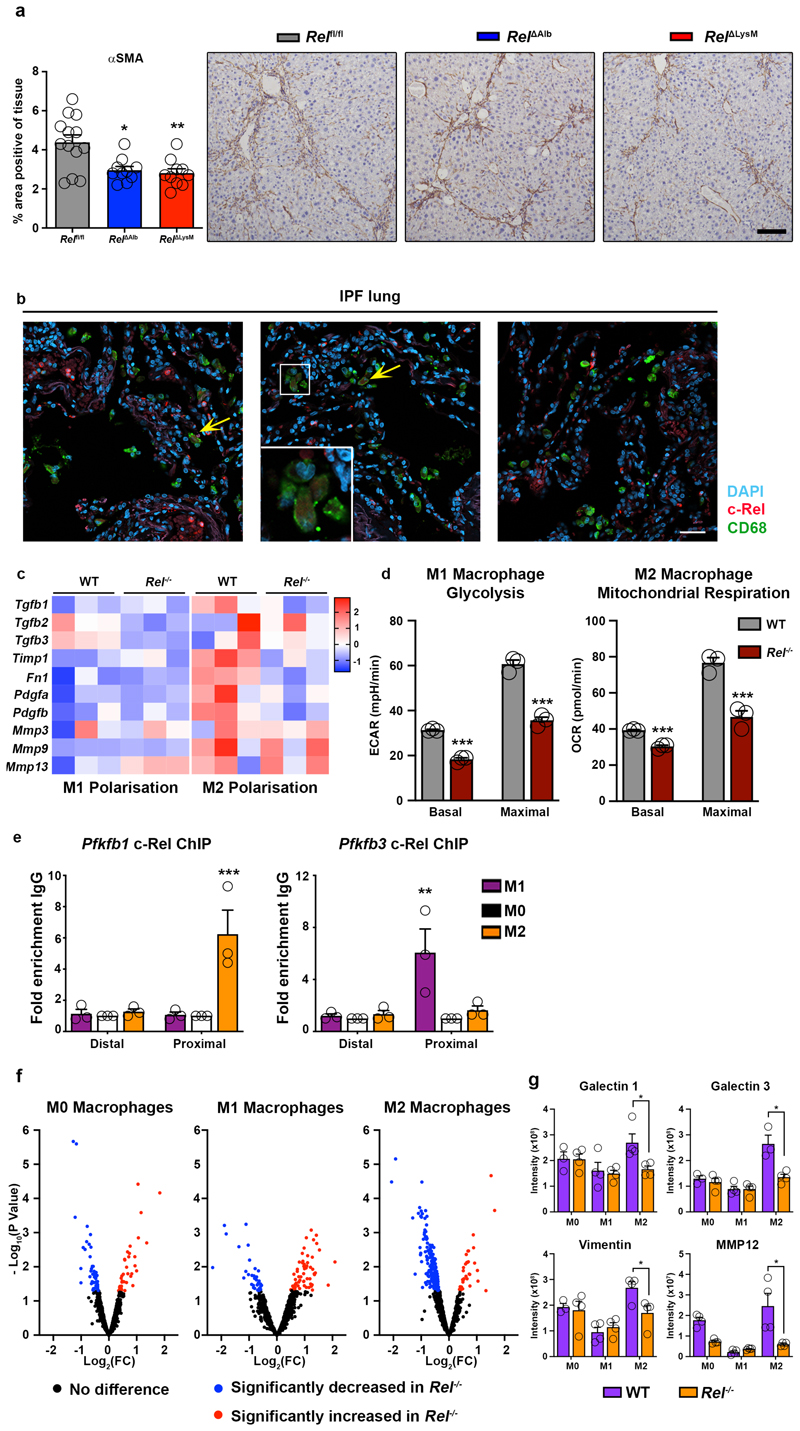
Extended Data Fig. 5. c-Rel regulates macrophages polarisation to drive tissue fibrosis.
(a) Histological assessment and representative images of αSMA stained liver sections in acute CCl4 injured in (n=13) Relflfl, (n=7) RelΔAlb (p=0.0116), and (n=10) Rel ΔLysM (p=0.0051) mice. Data are mean ± s.e.m. Scale bar equals 100 microns. P values were calculated using a one-way ANOVA with Tukey post- hoc t-test. (b) Representative low power immuno-fluorescence images show c-Rel (red), CD68 (green) and nuclear (blue) staining in diagnosed idiopathic pulmonary fibrosis lung sections. Yellow arrows denote co-localisaton of c-Rel and CD68. Scale bar equals 50 microns. Images are representative of n=8 IPF stained sections. (c) Heat map showing relative mRNA expression of fibrogenic markers; Tgfb1, Tgfb2, Tgfb3, TIMP1, Fn1, Pdgfa, Pdgfb, Mmp3, Mmp9 and Mmp13 in M1 and M2 polarised WT and Rel-/- BMDMs. (d) Graphs show, seahorse analysis of basal (p<0.0001) and maximal (p=0.0004) glycolysis (extracellular acidification rate, ECAR) and basal (p=0.0002) and maximal (p=0.0024) mitochondrial respiration (oxygen consumption rate, OCR) in M1 and M2 polarised WT and Rel-/- BMDMs. Data are mean ± s.e.m. from 3 independent cell isolations/group. (e) ChIP analysis of c-Rel binding to distal and proximal (p=0.004) regions of the Pfkfb3 promoter and to distal and proximal (p=0.003) regions of the Pfkfb1 promoters in WT BMDM in response to M1 and M2 polarisation. Data are mean ± s.e.m. from 3 independent cell isolations/group. (d-e) P values were calculated using two-way ANOVA with Tukey post-hoc t-test. Denoted significance refers to comparisons between WT and Rel-/- macrophages polarised to either an M1 or M2 phenotype. (f) Volcano plots show differentially expressed proteins detected by proteomic analysis of the secretome of M0, M1 and M2 polarised WT and Rel-/- BMDMs. (g) Graphs show relative levels of Galectin 1 (p=0.0031), Galectin 3 (p=0.0013), Vimentin (p=0.0128) and Matrix Metalloproteinase 12 (MMP12) (p<0.0001) expressed as Intensity x108 in M0, M1 and M2 polarised WT and Rel-/- BMDMs. Data are mean ± s.e.m. from 4 independent cell isolations/group generated 4 different donors/genotype. P values were calculated using the R package LIMMA (* P <0.05, ** P <0.01 and *** P <0.001).