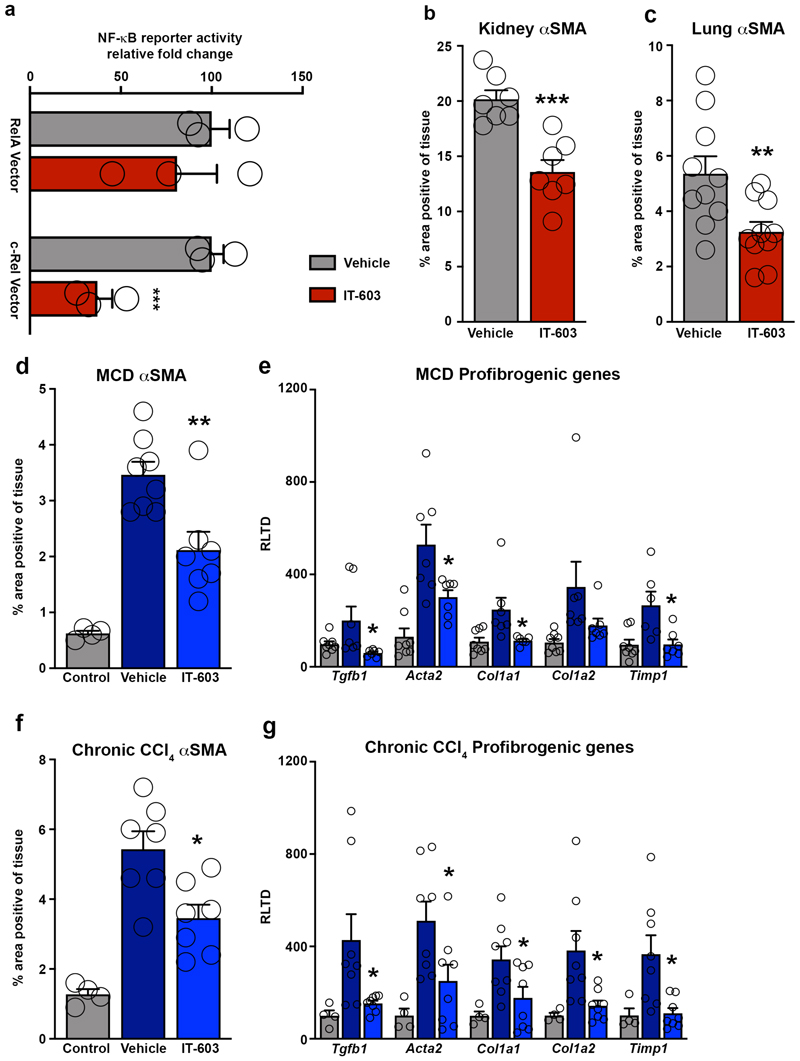
Extended Data Fig. 9. IT-603 attenuates fibrogenesis in murine models of fibrosis.
(a) U937 cells stably expressing 3xNF-(B-Luc reporter were transiently transfected with RelA or c-Rel expression plasmids. Graph shows RelA and c-Rel induced NF-κ(B luciferase reporter activity ± IT-603 therapy. Data are mean ± s.e.m. P value = 0.0039. P value was calculated using an unpaired two-sided t-test (***P<0.001) of 3 independent experiments. (b-c) Histological quantification of αSMA stained kidney (p=0.0004) or lungs (p=0.009) following their respective injury. Data are mean ± s.e.m. in 7 and 10 mice/group for kidney and lung respectively. P values were calculated using an unpaired two-sided t-test. (d) Histological quantification of αSMA stained chronic MCD diet injured livers at 2 weeks (pre-treatment) and 5 weeks ± therapeutic administration of IT-603 (p=0.0044). (e) Graphs showing relative hepatic expression of the fibrogenic genes; Tgfb1 (p=0.039), Acta2 (p=0.028), Col1a1 (p=0.036), Col1a2, and Timp1(p=0.015) in 2-week (pre-treatment) and 5-week methionine choline deficient diet (MCD) fed mice ± therapeutic administration of IT-603. Data are mean ± s.e.m. in n=4 control mice 8 vehicle and n=7 IT-603 treated mice/group. (f) Histological quantification of αSMA stained chronic CCl4 injured livers at 3 weeks (pre-treatment) or 8 weeks ± therapeutic administration of IT-603 (p=0.0105). (g) Graphs showing relative hepatic expression of the fibrogenic genes; Tgfb1 (p=0.029), Acta2 (p=0.032), Col1a1 (p=0.04), Col1a2 (p=0.016), and Timp1 (p=0.009) in chronic CCl4 injured livers at 3 week (pre-treatment), 8 week and ± therapeutic administration of IT-603 (from weeks 3-8). Data are mean ± s.e.m. in n=4 control mice 7 experimental mice/group. (c,e) P values were calculated using one-way ANOVA with Tukey post-hoc t-test. (d,f) P values were calculated using two-way ANOVA with Tukey post-hoc t-test (*P<0.05, ***P<0.001).