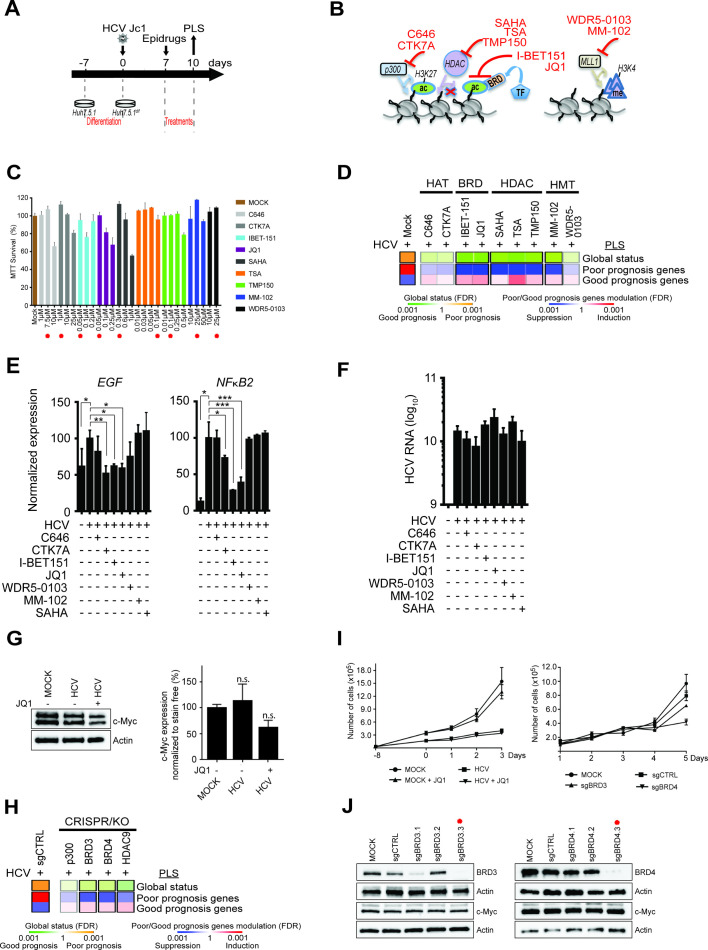
Figure 4.
Effect of small molecules and knockdown targeting chromatin modifiers and readers on gene expression driving hepatocellular carcinoma (HCC) risk in cell culture. (A) Schematic representation of the experimental setup. (B) Mode of action of inhibitors of chromatin modifiers/readers targeting histone acetylation and methylation. TF, transcription factors. (C) Cytotoxicity of the inhibitors in Huh7.5.1dif cells depicted in panel B. Cell viability of non-infected cells (n=3) was measured 72 hours following incubation with inhibitors using MTT assay. Results (mean±SEM) show percentage of viable cells in compound-treated conditions relative to untreated cells. The concentrations used for functional assays shown in panel D are indicated by red asterisks. (D) Effect of compounds on the prognostic liver signature (PLS) gene expression. Heatmaps show: (top) the classification of the PLS global status as poor (orange) or good (green) prognosis; (bottom) the significance of induction (red) or suppression (blue) of PLS poor-prognosis or good-prognosis genes. FDR, false discovery rate. (E) mRNA expression of EGF and NFκB2 in cells treated with the indicated compounds for 72 hours (*p<0.05; **p<0.01; ***p<0.001, unpaired t-test). (F) Inhibitors of chromatin modifiers/readers do not modulate HCV viral load in the experiment shown in panel E. (G) Western blot analysis showing c-Myc protein expression in the cell-based system. One representative gel is shown (see online supplementary figure S4). Right panel graph shows the quantification of western blot analysis intensities in arbitrary units normalised to total protein level (n=3) (stain-free staining). Results show the mean±SEM of integrated blot densities of four independent experiments and are not significantly (‘n.s.’) modulated. (H) Gene silencing of BRD3/4 and HDAC9 reverses the HCC high-risk status of the PLS in Huh7.5.1dif-Cas9 cells. HCV-infected Huh7.5.1dif-Cas9 cells expressing the PLS poor-prognosis status were transduced with lentiviral vectors coding for sgRNA targeting p300, BRD3, BRD4 and HDAC9 genes or non-targeting sgRNA (sgCTRL) (means of two experiments). (I) Proliferation analyses: (left) JQ1-treated or untreated Mock and HCV-infected cells; (right) Mock or HCV-infected Huh7.5.1dif-Cas9 cells transduced with lentiviral vectors coding for sgCTRL or sgRNA targeting BRD3 and BRD4 genes. (J) Analysis of BRD3, BRD4, c-Myc and Actin protein expression levels in Huh7.5.1dif-Cas9 cells KO for BRD3 or BRD4 by western blot analysis. Effective sgRNA for BRD3 and BRD4 genes used in the screening experiment shown in (H) are labelled with a red asterisk.