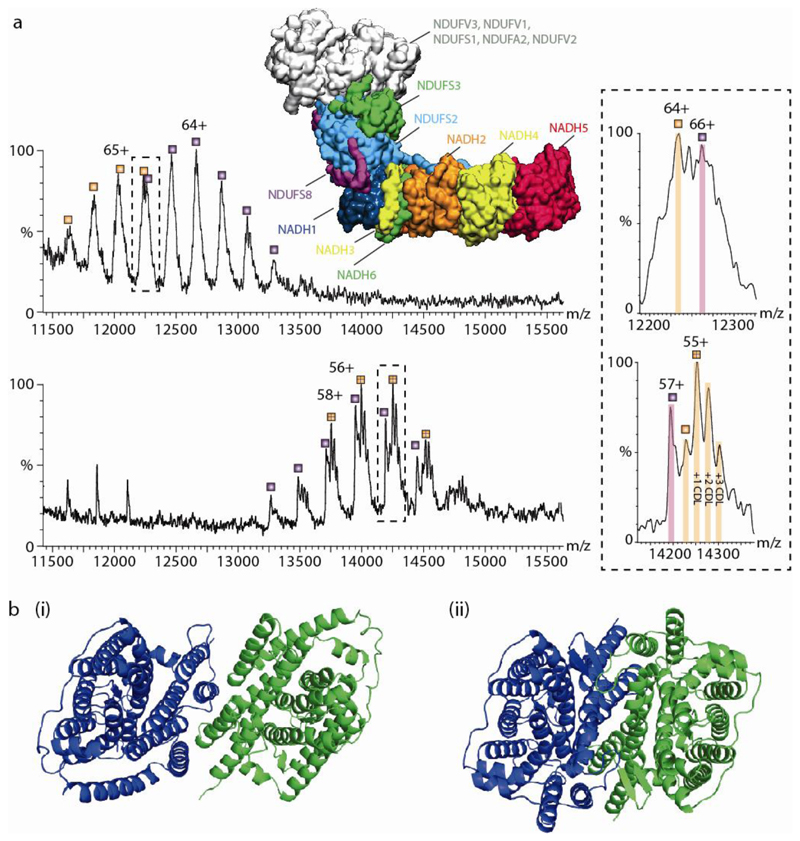
Fig. 3. The effect of detergents on lipid binding and structural integrity.
(a) Comparison of mass spectra of Complex I (CI) in detergent micelles (upper) and directly from vesicles (lower). Upper panel is mass spectrum recorded in a n-Dodecyl-β-D-Maltopyranoside (DDM) detergent micelle, which reveals two series of peaks (labeled orange and purple) differing in mass by 26,612 Da assigned to NDUFS3 (in green, within the protein model). The structure shown is from PDB:5LDW. Both series are assigned to the complete membrane region. The charge states differ between both conditions- the top panel is measured in 2x critical micellar concentration (CMC) of DDM, the bottom panel is CI lacking the N module ejected directly from native membranes with no recourse to detergents or other membrane mimetics. The lipid binding properties change in terms of both the extent and identity of bound lipids. However, the measured mass remains essentially the same for both the detergent extracted and SoLVe measurements (782,772 ± 35 Da compared to 782,478 ± 27 Da) for the membrane arm without NDUFS3 (orange series) and 809,269 ± 47 compared to 809,101 ± 57 Da for the membrane arm with NDUFS3 (purple series) respectively. (b) Comparison of UraA:Proton symporter in β-NG75, PDB: 3QE7 (i) whereby the dimer interface is deformed by the protrusion of the acyl chain of the detergent molecules between key helices in each monomer, and in fos-choline 9 and 1176 from PDB:5XLS (ii) showing a functional dimer.