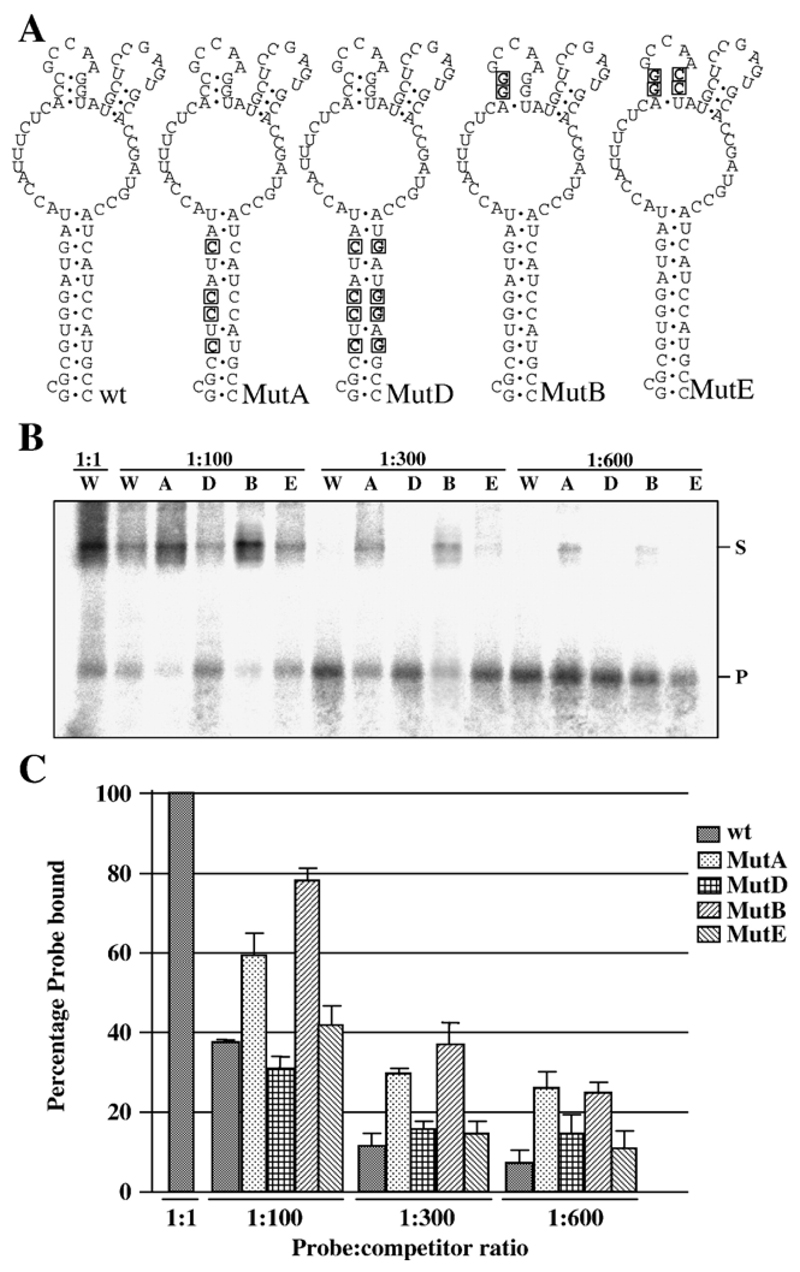
Fig. 1. Interaction between NS2 and mutant S10 RNAs.
(A) Mutations introduced into the wild-type (wt) sequence in BTV-10 S10 between nucleotides 99 and 169 are boxed and in bold. Changes were designed to destabilize the major stem (MutA) or a stem supporting a minor loop (MutB). Compensating mutations designed to allow the major stem (MutD) or minor stem (MutE) to reform were also produced. (B) EMSA data using purified NS2 and RNAs shown in panel A. All binding reactions were performed with 1 fmol probe and either 1:1, 1:100, 1:300 or 1:600 molar ratio of probe:competitor as indicated. W, A, D, B and E indicate that competitor RNA was wild type, MutA, MutD, MutB or MutE respectively. Position of free probe (P) and probe complexed with NS2 (S) are indicated. (C) Graph of combined data from three independent EMSA experiments. Error bars indicate the standard error of the mean. Note that second-site mutations that restore the RNA structure but not primary sequence show similar competition to wild-type RNA.