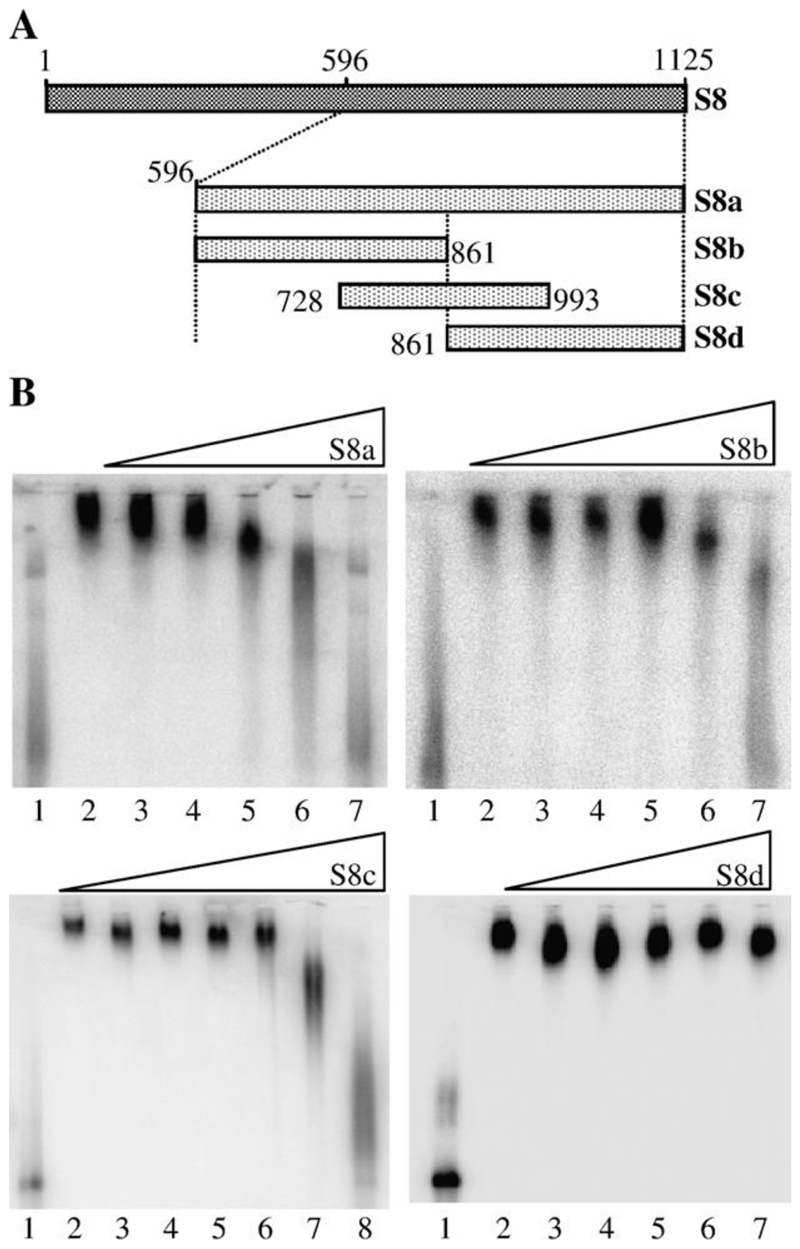
Fig. 2. Localization of the region of S8 RNA bound by NS2.
(A) Schematic of the truncations in S8 plus-strand RNA used as competitors in EMSA. Four fragments; S8a (nt 596–1125), S8b (nt 596–861), S8c (728–993) and S8d (861–1125) were used. (B) EMSA data. The S8 truncation used as competitor for full-length probe is shown above each panel. Lane 1 contains 1 × 10−9 M 32P- labeled full-length S8 probe only; lane 2, 32P-labeled probe and NS2. For S8a, S8b and S8d, lanes 3–7 contain 32P-labeled probe, NS2 and unlabeled competitor in a molar ratio of probe: competitor of 1:10, 1:100, 1:500, 1:1000 and 1:10,000, respectively. For S8c, lanes 3–8 contain 32P-labeled probe, NS2 and unlabeled competitor in a molar ratio of probe: competitor of 1:10, 1:100, 1:300, 1:600, 1:1000 and 1:10,000 respectively.