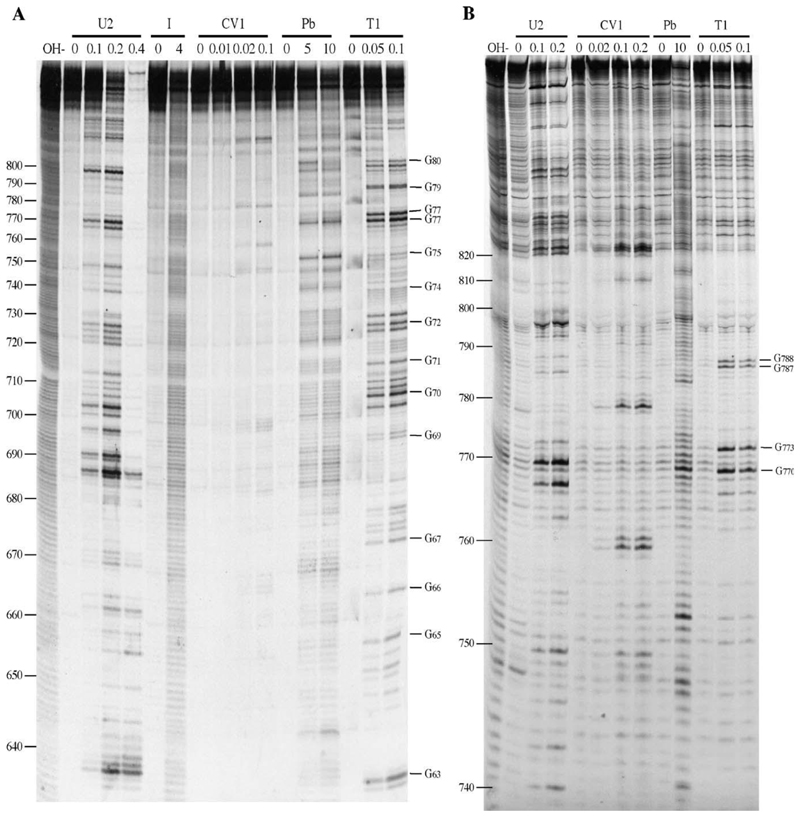
Fig. 5. Enzymatic and chemical structure probing of S8.
Base numbers correspond to those of the S8 cDNA. Products were analyzed on denaturing acrylamide/7M urea gels and data collected from 6%, 10% and 17.5% gels. Enzyme units per reaction (U2, T1), hours of reaction (I [imidazole]) and mM concentration in reaction (Pb [lead]) are shown. OH–, alkaline hydrolysis ladder; R, RNA only. (A) 6% denaturing acrylamide gel showing the results of chemical and enzymatic probing of S8b nt (nt 596–861). (B) As panel A, but structure mapping of S8c (nt 728–993) is shown. RNA structure probing analysis was also carried out on M6 and S9 RNA as detailed in the text