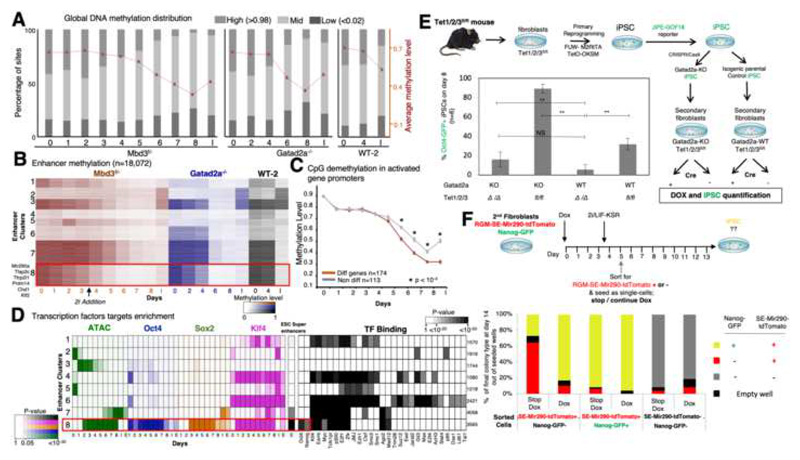
Fig. 3. Rapid DNA-demethylation of naïve ESC super-enhancers during conducive iPSC reprogramming.
A. Distribution of low (<0.02), mid (0.02-0.98) and high (>0.98) methylated CpG sites, along reprogramming. Average and SEM are indicated in red plot. B. Methylation level measured in covered enhancers (n=18,072), in Mbd3f/-, Gatad2a-/- and WT-2 systems. Enhancers are clustered into eight clusters using k-means. Cluster 8 consists of enhancers that undergo fast demethylation, compared to clusters 3 and 7. C. Average methylation measured in promoters of genes that were highly methylated (>80%) in day0. Genes that change their expression level (red) are compared to genes that do not change their expression level (gray). Wilcoxon p-value indicates places where methylation of differential genes is significantly lower than methylation of non-differential genes. D. Left: Enrichment of enhancer clusters, as shown in panel B, for OSK binding, DNA accessibility, and super enhancers, showing that cluster 8 is highly enriched for OSK binding and overlaps with super enhancers. Color shades represent FDR corrected enrichment p-value. Right: Enrichment of the same enhancer clusters to transcription factor binding, taken from hmChip database. Cluster size is indicated on the right. E. Experimental scheme summary. Reprogramming efficiency was measured by Oct4-GFP+ cells percentage in Tet1/2/3 null(Δ) and Tet1/2/3fl/fl with and without Gatad2a expression, after 8 days. **p<0.01, ***p<0.001 (Student’s t-test), n=6, error bars indicate SD. F. Secondary MEF harboring Mir290-RGM and Nanog GFP-reporter were sorted after reprogramming to 3 different populations: RGM-SE-Mir290-tdTomato positive cells (sorted at day 5), Nanog-GFP and Mir290-RGM positive cells (sorted at d10-14), and "double negative" cells (sorted at d5). The cells were seeded as single cell-per-well, and were treated with medium either supplemented with Dox or lacking Dox. On day 14 colonies were inspected for GFP and mCherry (RGM) markers.