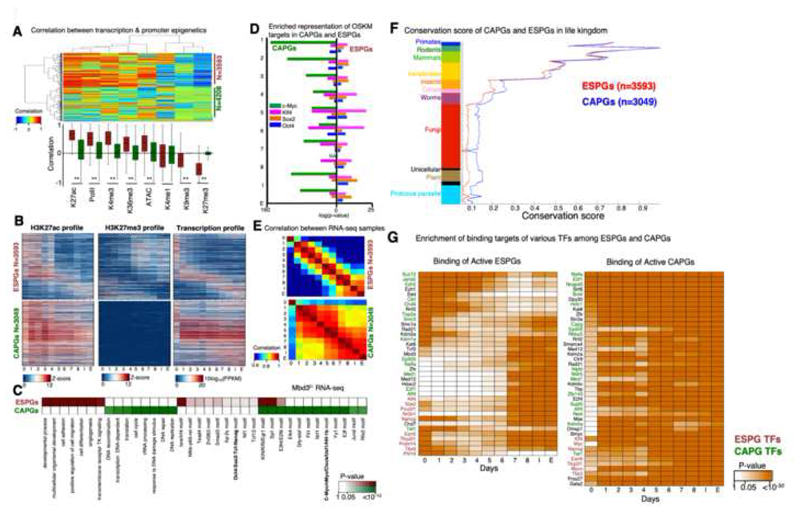
Fig. 4. Distinct regulation of cell fate genes and of biosynthetic processes by MYC.
A. Spearman correlation between the expression change of each differential gene (row) and change in promoter chromatin modification of each indicated mark (column). Analysis was done on all differential genes that have at least two marks in their promoter (i.e. with z-score>1 std), resulting in 7801 genes. Hierarchical clustering clustered the genes into two distinct groups: genes with correlation between gene expression and promoter epigenetic modifications (n=3593) and genes with no trend of correlation (n=4208). Top: clustered correlation matrix. Bottom: correlation distribution of each mark in the two gene groups, where red denote correlated group, and green denote the non-correlated group. **Wilcoxon p-value <10-170 B. Pattern of H3K27ac, H3K27me3 and expression in Epigenetically Switched Genes (ESPGs, top), and in genes with Consistency-Active-Promoters (CAPGs, bottom). Each row corresponds to a single gene, genes are sorted according to their expression pattern and the same sorting was applied to the epigenetic marks. C. Enrichment of GO categories and binding motifs in ESPGs (red), CAPGs (n=3049, green). Color shades indicate FDR corrected Fisher exact test p-value, or motif enrichment p-value. D. Enrichment of OSKM binding targets in promoters of ESPGs compared to CAPGs. Minus log10 of Fisher exact test p-values are indicated. E. Spearman correlation matrix between Mbd3f/- RNA-seq samples calculated over ESPGs (Top), showing a gradual change along reprogramming and over CAPGs (Bottom), showing two waves of change, on day 1, and on days 5-6. F. Conservation score of ESPGs and CAPGs calculated with Phylogene software74.Graph includes the mean and SEM values for each gene set (one tailed Wilcoxon test, p-value of nonvertebrate organisms <10-30). G. Scheme showing transcription factors that significantly bind ESPGs or CAPGs that are active in each day of reprogramming, based on ChIP-seq databases. Red – ESPGs transcription factors, Green – CAPGs factors. Orange shades – FDR corrected p-values (Fisher exact test).