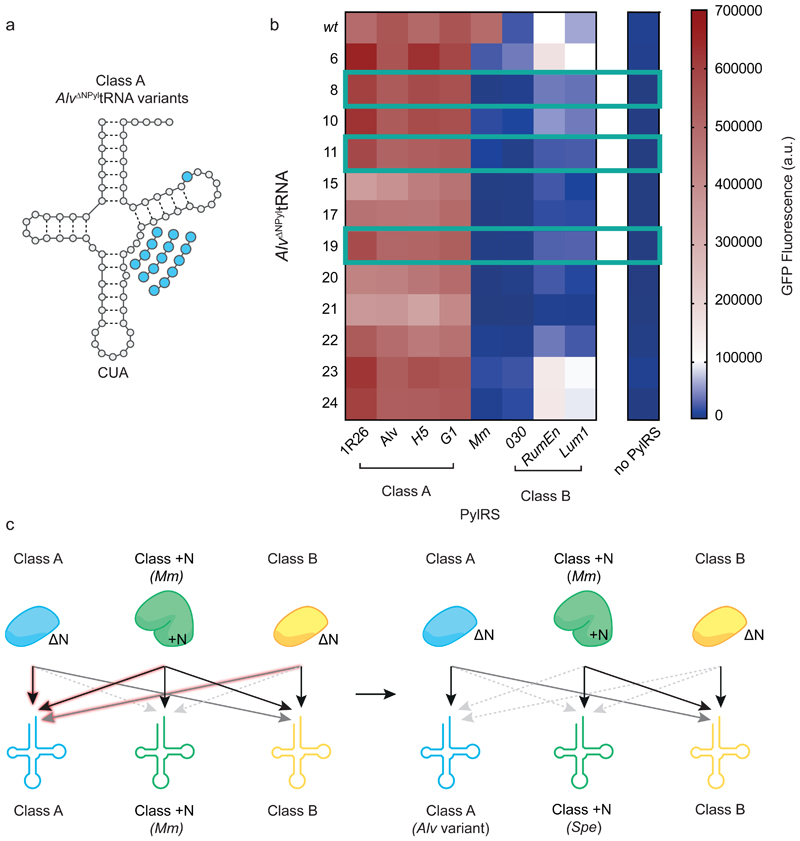
Figure 4. Identifying triply orthogonal and active Class A ΔNPyltRNAs.
a, Clover leaf structure of Alv ΔNPyltRNACUA variants with variable loop expansions and point mutations highlighted in blue. b, GFPHis6 fluorescence resulting from wild type Alv ΔNPyltRNACUA or the indicated Alv ΔNPyltRNACUA variant with each PylRS from cells containing GFP(150TAG)His6 in presence of BocK 1. Each heatmap value represents the average of three biological replicates. Bar charts including error bars showing s.d. are provided in Supplementary Fig. 13 and all numerical values are provided in Supplementary Table 3. Alv ΔNPyltRNA(8)CUA, Alv ΔNPyltRNA(11)CUA and Alv ΔNPyltRNA(19)CUA all showed improved orthogonality to MmPylRS and class B ΔNPylRSs while remaining active with class A ΔNPylRSs (highlighted with a green frame). c, Schematic of the progress made towards generating triply orthogonal PylRS/PyltRNA pairs when substituting wild-type Alv ΔNPyltRNA (left) with selected Alv ΔNPyltRNA variants (right). Black arrows indicate high activity, grey arrows indicate low activity and dashed grey arrows indicate minimal activity corresponding to functional orthogonality. The interactions of Alv ΔNPyltRNA that were manipulated in these experiments are highlighted.