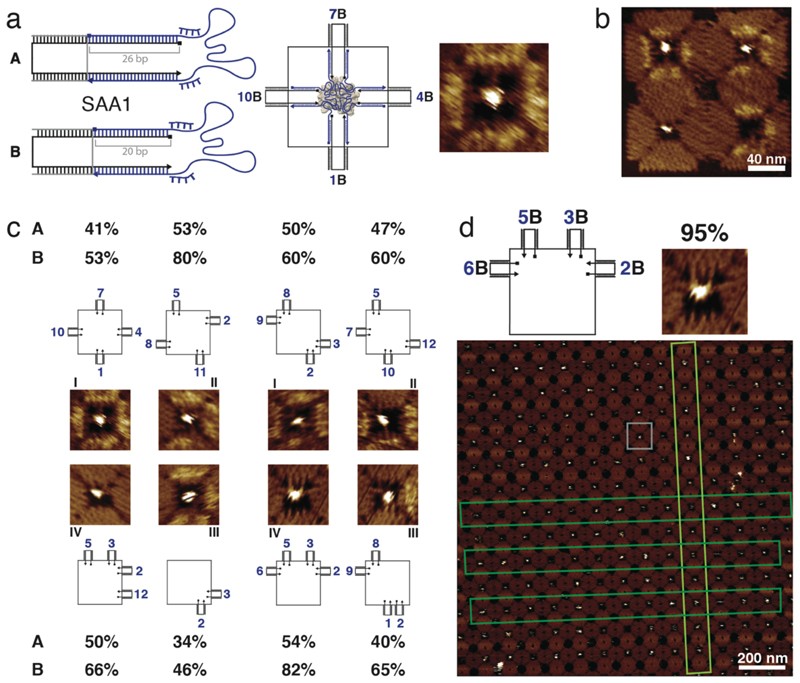
Figure 4. Binding of streptavidin to four identical aptamers in different configurations.
a, Scheme of the streptavidin aptamer SAA1 attached via spacers, each comprised of a 26 bp (case A) or a 20 bp stem (case B) and additional (dT)4 aptamer linkers. On the right, a 2D scheme and a zoomed-in AFM image of the (1,4,7,10) configuration for SAA1 in case B are shown. b, Example of an AFM image of a 2 x 2 array (barcode 3) including different configurations of four SAA1 aptamers in four cavities. c, AFM images and schemes for eight configurations of four SAA1 aptamers binding streptavidin. The binding yields for both long and short stems for these different configurations are given. Typical errors of the percentages are ≤ 3% (standard error of the mean, cf. Supplementary Fig. 15). d, Crystal formation of DNA origami structures with the best configuration (2B,3B,5B,6B) for binding to streptavidin (this configuration is shown in the scheme on top and the small examplary AFM image). In lattice shown in the large AFM image, the gray box highlights a single origami structure and the dark and light green boxes point out linear arrangements of structures within the crystal. In the context of the crystal, we observe a binding yield of ≈ 95%.