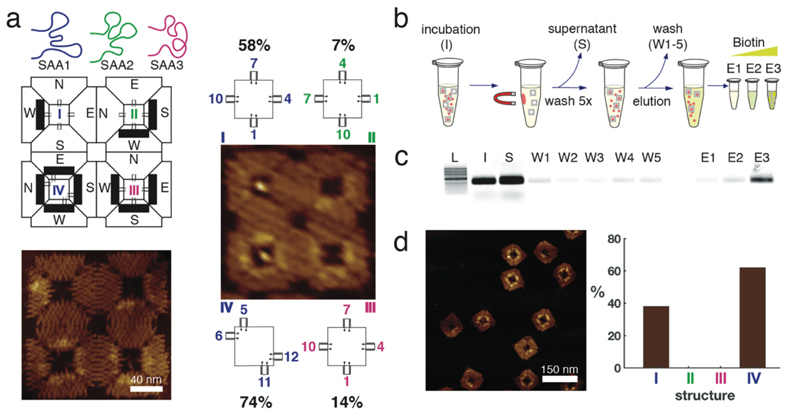
Figure 5. Evaluation of the enrichment for the nanostructures with the better binding configuration to streptavidin.
a, Schematic drawing of the three aptamers tested in the 2 x 2 array. Position I and IV have the SAA1 aptamer located in configurations (1,4,7,10) and (5,6,11,12), respectively. Position II and III have SAA2 and SAA3, respectively, in the symmetric configuration (1,4,7,10). AFM images of the array with (on the right) and without (bottom left) streptavidin. The percentages refer to the binding assay done together with different configurations in figure 3. b, scheme of the selection process of the higher affinity configuration using streptavidin coated magnetic particles. On the order of 109 microbeads were incubated with 20 nM nanostructures for 30 min at room temperature (I) and the supernatant (S) was removed using a magnetic separator. Subsequently, beads were washed five times with evolution buffer (W1-W5), and eluted with increasing concentrations of biotin (E1-E3). c, Agarose gel stained with SyBr Gold showing each step of the selection protocol. L, 1Kb ladder NEB, I, initial sample, S, supernatant, W1-W5, washing steps, E1-E3 elution with 0.8, 8 and 80 µM biotin respectively. d, AFM image of the biotin-eluted sample E3. The histogram shows the yield of the nanostructures purified from E3. Nanostructures in position II and III of the 2 x 2 array were not selected by this method. Percentages are derived from single AFM experiments to enable direct comparison under identical conditions (n=1, therefore no errors/error bars are given).