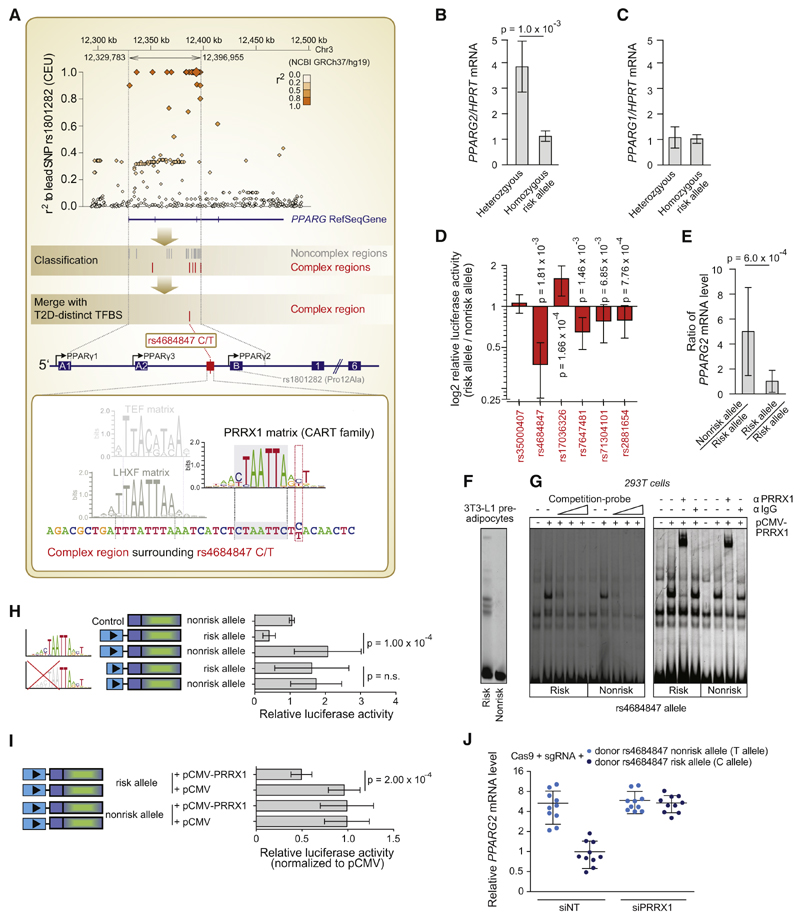
Figure 4. The Noncoding SNP rs4684847 by Binding the Homeobox Factor PRRX1, Represses PPARG2 Expression at the PPARG Diabetes Risk Locus.
(A) Top panel: an LD regional plot of the PPARG locus. Diamonds, tagSNP Pro12Ala and pairwise correlation of SNPs in LD (MAF ≥ 1%) against genomic position; blue, PPARG gene and exons. Middle/lower panel: classification of SNPs in complex regions (red lines) and noncomplex regions (gray lines) (PMCA steps 1–9, Figure 1A). Scanning of PPARG complex regions for T2D-distinct homeobox TFBS matrix families (CART, HOMF, HBOX, NKX6, BCDF, PDX1; Figure 3B) pinpoints rs4684847 (C/T), based on its overlap with the CART binding matrix for PRRX1 (step 10, Figure 1A). Zoom-in, human PPARG gene; arrows, transcription start site (TSS) of PPARG1-3 mRNA isoforms; boxes, coding exons (filled) and untranslated exons (open); lines, introns. Second zoom-in, CRM at rs4684847; the PRRX1 matrix co-occurs with diverse TFBS matrices in consistent orientation and distance range across species, exemplarily illustrated by one conserved TFBS module (UTFBS_in_modules = 3; TFBS matrices: PRRX1, TEF, LHXF).
(B and C) Genotype-dependent mRNA expression in undifferentiated hASCs genotyped for Pro12Ala and rs4684847 (r2 = 1.0). qPCR of PPARG1 and PPARG2 mRNA isoforms (standardized to HPRT) homozygous CC risk (n = 9) and CT nonrisk allele carriers (n = 5) normalized to mean for CC. Mean ± SD, t test.
(D) Validation of cis-regulatory predictions for complex regions at the PPARG locus. Quantified change in reporter activity in 3T3-L1 adipocytes is shown for each SNP, using luciferase constructs harboring the risk or nonrisk alleles, representing an activating or repressing effect of the risk allele on transcriptional activity.
Mean ± SD, n = 3–14, paired t test.
(E) Allele-specific primer extension analysis in hASCs of heterozygous rs4684847 carriers (n = 6) normalized to mean risk allele levels (D). Mean ± SD, Mann-Whitney U test.
(F and G) Increased PRRX1 binding at the risk allele in EMSAs with rs4684847 allelic probes and 3T3-L1 preadipocyte nuclear extracts (F), confirmed by competition with cold PRRX1 probe (G, left panel) and PRRX1 antibody shift of protein-DNA complex in 293T with ectopically expressed PRRX1 (G, right panel).
(H) Reporter assays with constructs harboring the rs4684847 risk and nonrisk allele in 3T3-L1 preadipocytes. Truncation of the PRRX1 matrix without affecting rs4684847 reveals abrogated allelic cis-regulatory activity. Mean ± SD, n = 9, paired t test.
(I) Inhibition of reporter activity (normalized to pCMV control) at the rs4684847 risk allele by ectopic expression of PRRX1 in 3T3-L1 preadipocytes. Mean ± SD; n = 9, paired t test.
(J) Regulation of PPARG2 mRNA expression in SGBS adipocytes with the CC risk allele, or TT nonrisk allele introduced by CRISPR/Cas9 genome editing approach. siPRRX1 and siNT transfection concurrent with induction of differentiation, PPARG2 mRNA assessed by quantitative RT-PCR (qRT-PCR), standardized to HPRT. Mean ± SD, n = 12, t test. siNT, nontargeting siRNA.